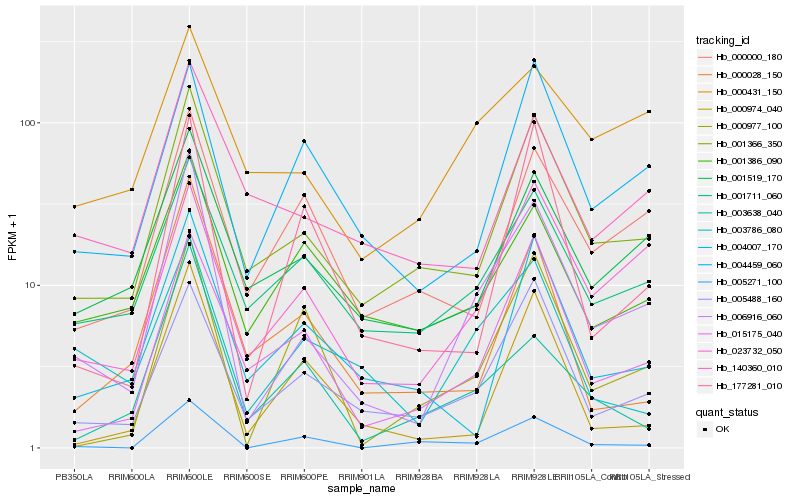
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006916_060 |
0.0 |
transcription factor |
TF Family: MIKC |
PREDICTED: truncated transcription factor CAULIFLOWER A-like [Jatropha curcas] |
| 2 |
Hb_001366_350 |
0.1791101624 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase isoform X2 [Jatropha curcas] |
| 3 |
Hb_005488_160 |
0.2067640376 |
- |
- |
PREDICTED: maltose excess protein 1, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_015175_040 |
0.2112330832 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001519_170 |
0.2124960419 |
- |
- |
PREDICTED: K(+) efflux antiporter 3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_023732_050 |
0.2173712468 |
- |
- |
aldo-keto reductase, putative [Ricinus communis] |
| 7 |
Hb_005271_100 |
0.219119155 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_001711_060 |
0.2240519538 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 9 |
Hb_003638_040 |
0.226450518 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 10 |
Hb_000977_100 |
0.2286003688 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: acetolactate synthase small subunit 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003786_080 |
0.2294724462 |
- |
- |
electron transporter, putative [Ricinus communis] |
| 12 |
Hb_000028_150 |
0.2298586986 |
transcription factor |
TF Family: Tify |
PREDICTED: protein TIFY 6B isoform X1 [Jatropha curcas] |
| 13 |
Hb_000974_040 |
0.233345941 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Jatropha curcas] |
| 14 |
Hb_000431_150 |
0.2335655031 |
- |
- |
ribosomal protein L27, putative [Ricinus communis] |
| 15 |
Hb_177281_010 |
0.2343624194 |
- |
- |
PREDICTED: 50S ribosomal protein 5, chloroplastic-like [Citrus sinensis] |
| 16 |
Hb_140360_010 |
0.2351996365 |
- |
- |
PREDICTED: 50S ribosomal protein L11, chloroplastic-like [Populus euphratica] |
| 17 |
Hb_004459_060 |
0.2370233897 |
- |
- |
50S ribosomal protein L34, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_004007_170 |
0.2372500828 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 19 |
Hb_001386_090 |
0.2384644602 |
- |
- |
PREDICTED: uncharacterized protein LOC105632529 [Jatropha curcas] |
| 20 |
Hb_000000_180 |
0.2398717218 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |