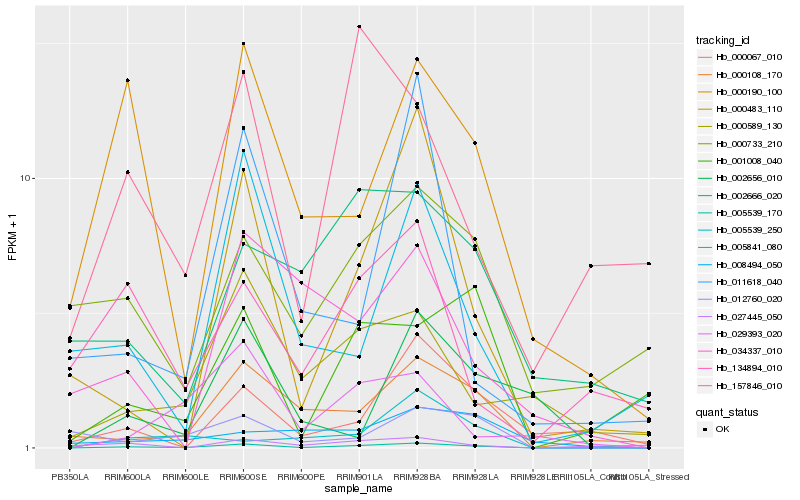
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005539_170 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC101502180 [Cicer arietinum] |
| 2 |
Hb_000108_170 |
0.2557183596 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |
| 3 |
Hb_000483_110 |
0.2769416848 |
transcription factor |
TF Family: zf-HD |
Mini zinc finger 2 isoform 2, partial [Theobroma cacao] |
| 4 |
Hb_000190_100 |
0.280485003 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0002s04880g [Populus trichocarpa] |
| 5 |
Hb_000589_130 |
0.291418207 |
desease resistance |
Gene Name: NB-ARC |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_001008_040 |
0.2933608748 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 7 |
Hb_029393_020 |
0.2993926295 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 12-like [Jatropha curcas] |
| 8 |
Hb_000067_010 |
0.3007885847 |
- |
- |
hypothetical protein B456_002G147500 [Gossypium raimondii] |
| 9 |
Hb_012760_020 |
0.3070684509 |
- |
- |
PREDICTED: uncharacterized protein LOC105647080 [Jatropha curcas] |
| 10 |
Hb_005841_080 |
0.3073549055 |
- |
- |
hypothetical protein POPTR_0018s11370g [Populus trichocarpa] |
| 11 |
Hb_000733_210 |
0.3073710592 |
- |
- |
putative serine/threonine protein phosphatase [Angiostrongylus costaricensis] |
| 12 |
Hb_005539_250 |
0.3145232752 |
- |
- |
- |
| 13 |
Hb_002666_020 |
0.314541259 |
- |
- |
VAMP/SYNAPTOBREVIN-ASSOCIATED protein 27-1 [Populus trichocarpa] |
| 14 |
Hb_134894_010 |
0.3177103146 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL5-like isoform X2 [Gossypium raimondii] |
| 15 |
Hb_034337_010 |
0.3211924487 |
- |
- |
vesicle-associated membrane protein, putative [Ricinus communis] |
| 16 |
Hb_002656_010 |
0.322587687 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_157846_010 |
0.3228439761 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 18 |
Hb_008494_050 |
0.3262210395 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 19 |
Hb_011618_040 |
0.3281328658 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_027445_050 |
0.3323833458 |
- |
- |
PREDICTED: titin isoform X13 [Jatropha curcas] |