| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
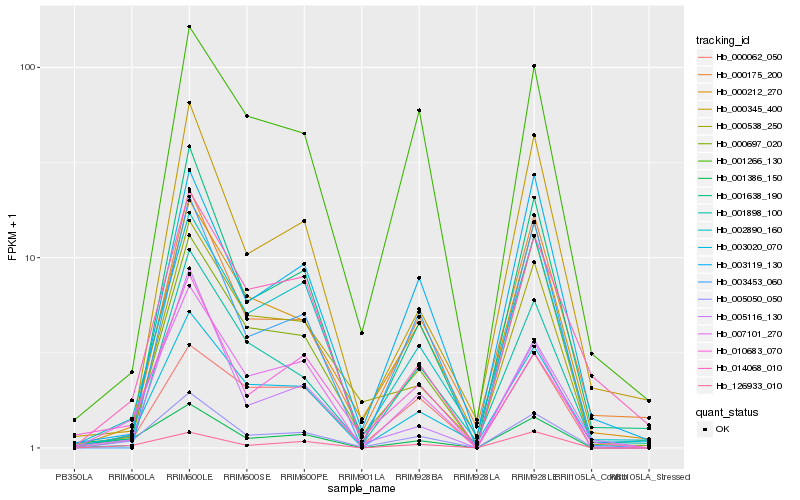
Hb_005050_050 |
0.0 |
- |
- |
PREDICTED: wall-associated receptor kinase 5-like [Glycine max] |
| 2 |
Hb_007101_270 |
0.1148369044 |
- |
- |
PREDICTED: uncharacterized protein LOC105640496 [Jatropha curcas] |
| 3 |
Hb_003020_070 |
0.1345143853 |
- |
- |
PREDICTED: uncharacterized protein LOC103997323 [Musa acuminata subsp. malaccensis] |
| 4 |
Hb_001386_150 |
0.140234745 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 5 |
Hb_000212_270 |
0.141231265 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 6 |
Hb_002890_160 |
0.1425545961 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X1 [Jatropha curcas] |
| 7 |
Hb_010683_070 |
0.1497980097 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase CMT2 isoform X1 [Jatropha curcas] |
| 8 |
Hb_014068_010 |
0.1512247292 |
- |
- |
PREDICTED: phototropin-1 [Jatropha curcas] |
| 9 |
Hb_126933_010 |
0.1554445297 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 10 |
Hb_001638_190 |
0.1567011893 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_000175_200 |
0.1588028009 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_003119_130 |
0.1680568222 |
- |
- |
hypothetical protein POPTR_0017s07770g [Populus trichocarpa] |
| 13 |
Hb_001898_100 |
0.1694814507 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 14 |
Hb_000697_020 |
0.1700896342 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 15 |
Hb_000538_250 |
0.1720155248 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g07650 [Jatropha curcas] |
| 16 |
Hb_001266_130 |
0.1720267905 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase-like [Jatropha curcas] |
| 17 |
Hb_000062_050 |
0.1721970402 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_005116_130 |
0.1736420532 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000345_400 |
0.1744932407 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_003453_060 |
0.1751655424 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |