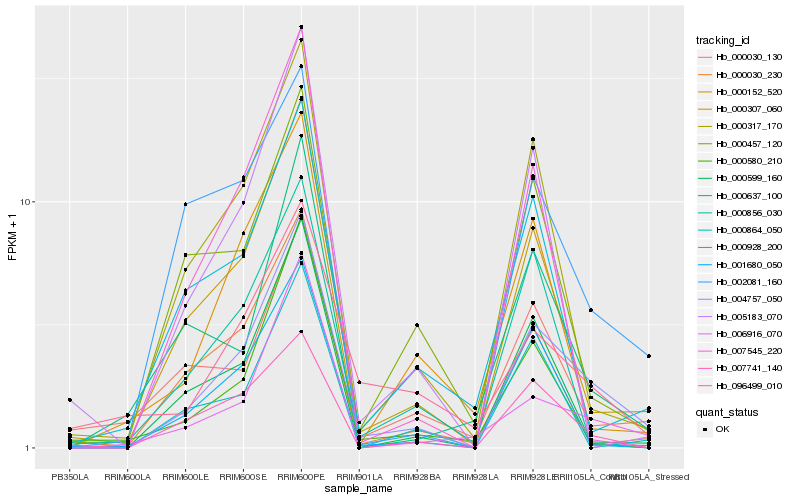
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004757_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_000307_060 |
0.148993109 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 3 |
Hb_006916_070 |
0.1795014178 |
transcription factor |
TF Family: MIKC |
PREDICTED: developmental protein SEPALLATA 1-like [Jatropha curcas] |
| 4 |
Hb_000599_160 |
0.1837085833 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 21/22 [Jatropha curcas] |
| 5 |
Hb_000317_170 |
0.1842730653 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 6 |
Hb_007741_140 |
0.1926591582 |
- |
- |
PREDICTED: SPX domain-containing protein 3-like isoform X2 [Populus euphratica] |
| 7 |
Hb_002081_160 |
0.1968398812 |
- |
- |
hypothetical protein OsI_35059 [Oryza sativa Indica Group] |
| 8 |
Hb_000030_230 |
0.2027005213 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 9 |
Hb_000928_200 |
0.2065850132 |
- |
- |
PREDICTED: RING-H2 finger protein ATL52-like [Jatropha curcas] |
| 10 |
Hb_005183_070 |
0.2066728236 |
- |
- |
PREDICTED: calmodulin-like protein 5 [Solanum lycopersicum] |
| 11 |
Hb_001680_050 |
0.2067538988 |
- |
- |
- |
| 12 |
Hb_000457_120 |
0.2074259581 |
- |
- |
PREDICTED: death-associated protein kinase dapk-1 [Jatropha curcas] |
| 13 |
Hb_000152_520 |
0.208049373 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0015s14600g [Populus trichocarpa] |
| 14 |
Hb_000637_100 |
0.2099374953 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26-like [Jatropha curcas] |
| 15 |
Hb_007545_220 |
0.2101706 |
- |
- |
PREDICTED: uncharacterized protein LOC105632565 [Jatropha curcas] |
| 16 |
Hb_096499_010 |
0.2130740857 |
- |
- |
hypothetical protein JCGZ_14833 [Jatropha curcas] |
| 17 |
Hb_000030_130 |
0.2132981818 |
transcription factor |
TF Family: G2-like |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_000580_210 |
0.2132990988 |
- |
- |
PREDICTED: cysteine-rich repeat secretory protein 15 [Jatropha curcas] |
| 19 |
Hb_000864_050 |
0.2145341287 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_000856_030 |
0.2154458329 |
- |
- |
ATP binding protein, putative [Ricinus communis] |