| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
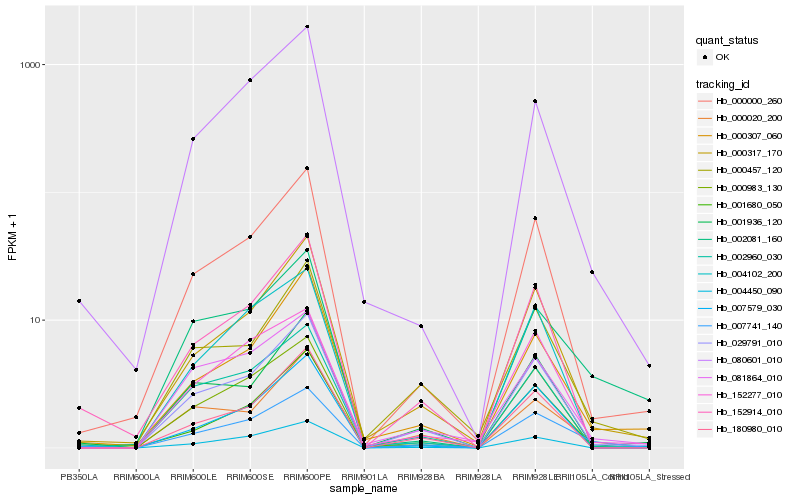
Hb_007741_140 |
0.0 |
- |
- |
PREDICTED: SPX domain-containing protein 3-like isoform X2 [Populus euphratica] |
| 2 |
Hb_000317_170 |
0.1261488335 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RING1 [Jatropha curcas] |
| 3 |
Hb_029791_010 |
0.1272493559 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 4 |
Hb_000000_260 |
0.1309940639 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 1 [Jatropha curcas] |
| 5 |
Hb_152914_010 |
0.1388897096 |
- |
- |
PREDICTED: polcalcin Syr v 3-like [Nicotiana sylvestris] |
| 6 |
Hb_001680_050 |
0.143134127 |
- |
- |
- |
| 7 |
Hb_000983_130 |
0.1457887866 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 8 |
Hb_080601_010 |
0.1484983043 |
- |
- |
metallothionein 3-like protein [Hevea brasiliensis] |
| 9 |
Hb_002960_030 |
0.1498263498 |
- |
- |
hypothetical protein POPTR_0002s02190g [Populus trichocarpa] |
| 10 |
Hb_004450_090 |
0.1503438716 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 11 |
Hb_007579_030 |
0.1510587298 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 12 |
Hb_004102_200 |
0.1513455143 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 13 |
Hb_152277_010 |
0.1521351983 |
- |
- |
PREDICTED: 9-cis-epoxycarotenoid dioxygenase NCED3, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_000457_120 |
0.1530227449 |
- |
- |
PREDICTED: death-associated protein kinase dapk-1 [Jatropha curcas] |
| 15 |
Hb_000020_200 |
0.1549855267 |
- |
- |
hypothetical protein EUGRSUZ_B00411 [Eucalyptus grandis] |
| 16 |
Hb_081864_010 |
0.1551231447 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000307_060 |
0.1561554691 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 18 |
Hb_002081_160 |
0.1588372237 |
- |
- |
hypothetical protein OsI_35059 [Oryza sativa Indica Group] |
| 19 |
Hb_180980_010 |
0.1588910615 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Malus domestica] |
| 20 |
Hb_001936_120 |
0.1595780483 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |