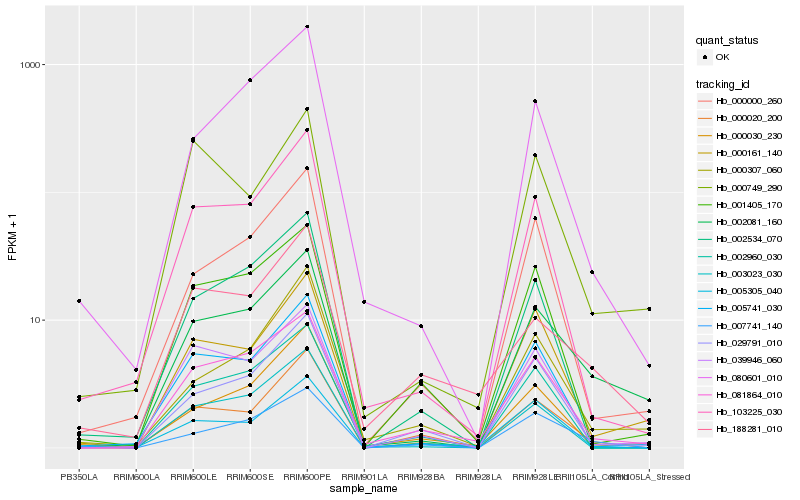
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002081_160 |
0.0 |
- |
- |
hypothetical protein OsI_35059 [Oryza sativa Indica Group] |
| 2 |
Hb_000161_140 |
0.145845883 |
- |
- |
PREDICTED: WEB family protein At3g51220 [Jatropha curcas] |
| 3 |
Hb_081864_010 |
0.1513231724 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_007741_140 |
0.1588372237 |
- |
- |
PREDICTED: SPX domain-containing protein 3-like isoform X2 [Populus euphratica] |
| 5 |
Hb_039946_060 |
0.1646710086 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor TINY-like [Jatropha curcas] |
| 6 |
Hb_103225_030 |
0.1654677742 |
transcription factor |
TF Family: Orphans |
DNA binding protein, putative [Ricinus communis] |
| 7 |
Hb_000000_260 |
0.1654732523 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 1 [Jatropha curcas] |
| 8 |
Hb_080601_010 |
0.1678442186 |
- |
- |
metallothionein 3-like protein [Hevea brasiliensis] |
| 9 |
Hb_005741_030 |
0.1714779081 |
- |
- |
PREDICTED: PI-PLC X domain-containing protein At5g67130-like [Jatropha curcas] |
| 10 |
Hb_000307_060 |
0.1724794906 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 8-like [Jatropha curcas] |
| 11 |
Hb_005305_040 |
0.1738895949 |
transcription factor |
TF Family: C3H |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_029791_010 |
0.1745324502 |
- |
- |
hypothetical protein POPTR_0014s17480g [Populus trichocarpa] |
| 13 |
Hb_000020_200 |
0.1784757392 |
- |
- |
hypothetical protein EUGRSUZ_B00411 [Eucalyptus grandis] |
| 14 |
Hb_000749_290 |
0.1787782847 |
- |
- |
PREDICTED: uncharacterized protein LOC105644242 [Jatropha curcas] |
| 15 |
Hb_001405_170 |
0.1793013101 |
- |
- |
aspartyl protease family protein [Populus trichocarpa] |
| 16 |
Hb_000030_230 |
0.1800163629 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 17 |
Hb_003023_030 |
0.1825255655 |
- |
- |
PREDICTED: MATE efflux family protein 5-like isoform X1 [Citrus sinensis] |
| 18 |
Hb_002534_070 |
0.182782192 |
- |
- |
PREDICTED: uncharacterized protein LOC105640545 [Jatropha curcas] |
| 19 |
Hb_188281_010 |
0.1843542023 |
- |
- |
carboxylic ester hydrolase, putative [Ricinus communis] |
| 20 |
Hb_002960_030 |
0.1849764098 |
- |
- |
hypothetical protein POPTR_0002s02190g [Populus trichocarpa] |