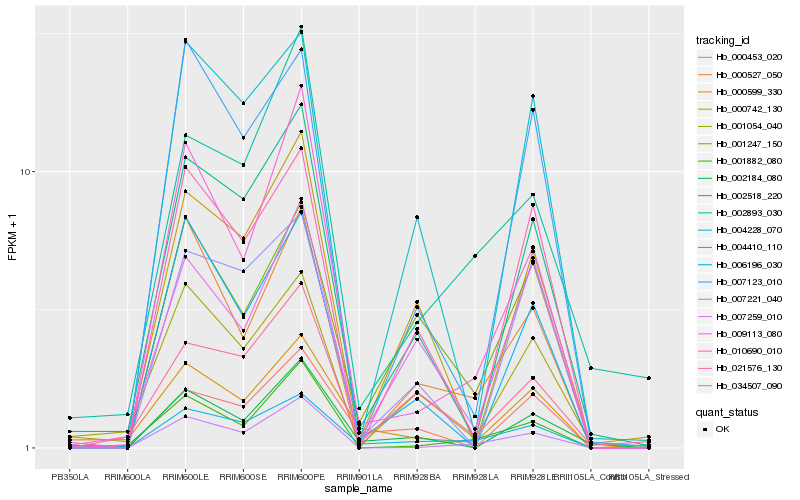
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004410_110 |
0.0 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 2 |
Hb_001247_150 |
0.1441229023 |
- |
- |
hypothetical protein JCGZ_23179 [Jatropha curcas] |
| 3 |
Hb_000527_050 |
0.1514977863 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_010690_010 |
0.1596977638 |
- |
- |
PREDICTED: NADPH-dependent 1-acyldihydroxyacetone phosphate reductase [Jatropha curcas] |
| 5 |
Hb_002518_220 |
0.1623342027 |
- |
- |
PREDICTED: uncharacterized protein LOC105631001 isoform X2 [Jatropha curcas] |
| 6 |
Hb_007259_010 |
0.1693058146 |
- |
- |
hypothetical protein CISIN_1g0073972mg, partial [Citrus sinensis] |
| 7 |
Hb_034507_090 |
0.1763848356 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 8 |
Hb_002893_030 |
0.1775228551 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Populus euphratica] |
| 9 |
Hb_000599_330 |
0.1805781645 |
- |
- |
hypothetical protein JCGZ_17661 [Jatropha curcas] |
| 10 |
Hb_021576_130 |
0.1809268495 |
- |
- |
PREDICTED: chorismate mutase 2 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002184_080 |
0.1814871474 |
- |
- |
triacylglycerol lipase, putative [Ricinus communis] |
| 12 |
Hb_000453_020 |
0.1817563085 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |
| 13 |
Hb_007123_010 |
0.1818845799 |
- |
- |
PREDICTED: auxin efflux carrier component 3-like [Jatropha curcas] |
| 14 |
Hb_007221_040 |
0.1819527856 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g24010 isoform X2 [Populus euphratica] |
| 15 |
Hb_006196_030 |
0.1820726685 |
- |
- |
PREDICTED: uncharacterized protein LOC105650679 [Jatropha curcas] |
| 16 |
Hb_004228_070 |
0.1846409169 |
- |
- |
PREDICTED: uncharacterized protein LOC103338699 [Prunus mume] |
| 17 |
Hb_009113_080 |
0.1863523247 |
- |
- |
PREDICTED: deoxynucleoside triphosphate triphosphohydrolase SAMHD1 homolog [Jatropha curcas] |
| 18 |
Hb_000742_130 |
0.1868870012 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g30570 [Jatropha curcas] |
| 19 |
Hb_001054_040 |
0.1887867177 |
- |
- |
hypothetical protein JCGZ_25310 [Jatropha curcas] |
| 20 |
Hb_001882_080 |
0.1887989625 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC103324347 [Prunus mume] |