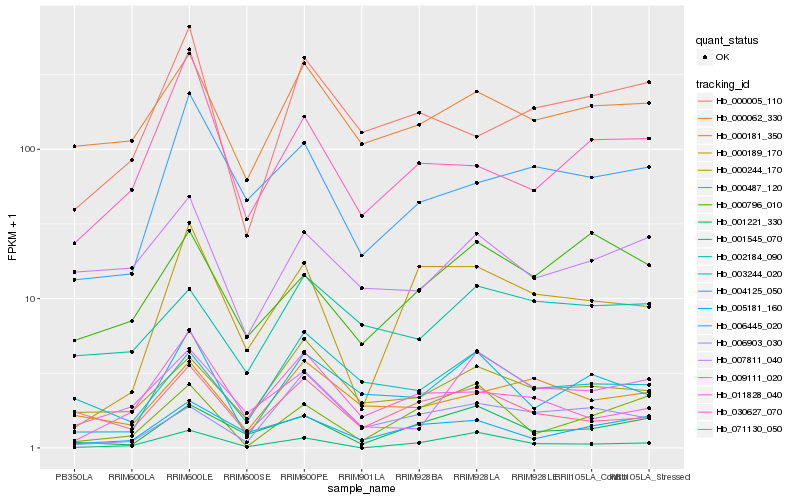
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004125_050 |
0.0 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 2 |
Hb_003244_020 |
0.1724928248 |
- |
- |
PREDICTED: serine carboxypeptidase-like 27 [Jatropha curcas] |
| 3 |
Hb_000244_170 |
0.1832191437 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK2 [Jatropha curcas] |
| 4 |
Hb_000062_330 |
0.1832221042 |
- |
- |
unknown [Lotus japonicus] |
| 5 |
Hb_071130_050 |
0.1967505273 |
- |
- |
PREDICTED: intracellular protein transport protein USO1 [Jatropha curcas] |
| 6 |
Hb_000005_110 |
0.2026967058 |
- |
- |
histone H2B1 [Hevea brasiliensis] |
| 7 |
Hb_005181_160 |
0.2054768414 |
- |
- |
PREDICTED: uncharacterized protein LOC105643546 [Jatropha curcas] |
| 8 |
Hb_001221_330 |
0.2104387457 |
- |
- |
PREDICTED: uncharacterized protein LOC105648791 [Jatropha curcas] |
| 9 |
Hb_000487_120 |
0.2108793128 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002184_090 |
0.2128512989 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_000181_350 |
0.2140603221 |
- |
- |
PREDICTED: uncharacterized protein LOC105111090 [Populus euphratica] |
| 12 |
Hb_030627_070 |
0.2150437017 |
- |
- |
histone H4 [Zea mays] |
| 13 |
Hb_007811_040 |
0.2151076865 |
- |
- |
PREDICTED: trafficking protein particle complex subunit 4 isoform X1 [Gossypium raimondii] |
| 14 |
Hb_006903_030 |
0.2179481943 |
- |
- |
PREDICTED: uncharacterized protein LOC105640666 [Jatropha curcas] |
| 15 |
Hb_000796_010 |
0.2187455423 |
- |
- |
PREDICTED: nitrogen regulatory protein P-II homolog [Jatropha curcas] |
| 16 |
Hb_000189_170 |
0.220040733 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 17 |
Hb_009111_020 |
0.2217957383 |
- |
- |
PREDICTED: calcium-dependent protein kinase 20-like [Jatropha curcas] |
| 18 |
Hb_011828_040 |
0.2224170263 |
- |
- |
PREDICTED: uncharacterized protein LOC105642173 isoform X2 [Jatropha curcas] |
| 19 |
Hb_001545_070 |
0.2243363719 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006445_020 |
0.225104002 |
- |
- |
hypothetical protein L484_026741 [Morus notabilis] |