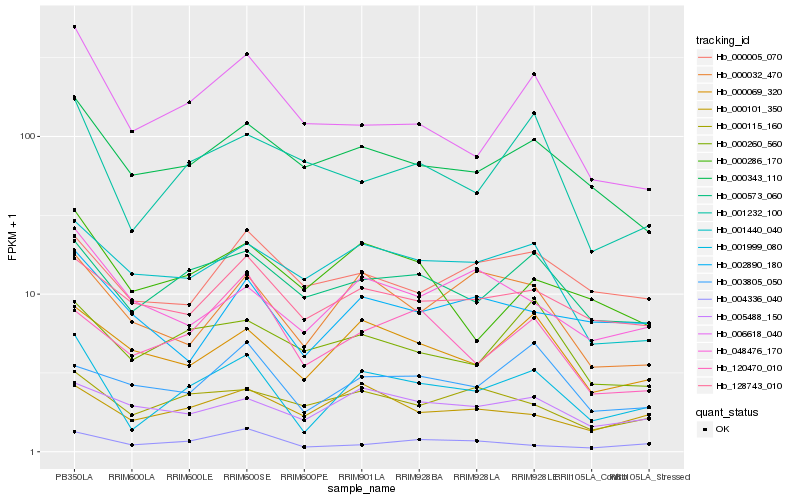
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001999_080 |
0.0 |
- |
- |
CCCH-type zinc finger protein [Hevea brasiliensis] |
| 2 |
Hb_000573_060 |
0.1646294756 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2D isoform X1 [Jatropha curcas] |
| 3 |
Hb_000069_320 |
0.1710116631 |
- |
- |
PREDICTED: uncharacterized protein LOC105642846 [Jatropha curcas] |
| 4 |
Hb_000343_110 |
0.1749084389 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 5 |
Hb_000032_470 |
0.1789851772 |
- |
- |
PREDICTED: uncharacterized protein LOC103415826 [Malus domestica] |
| 6 |
Hb_006618_040 |
0.1817701709 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 7 |
Hb_001232_100 |
0.1830394146 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 8 |
Hb_000101_350 |
0.1834081854 |
desease resistance |
Gene Name: NB-ARC |
NB-ARC domain-containing disease resistance protein, putative [Theobroma cacao] |
| 9 |
Hb_128743_010 |
0.1852492518 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 10 |
Hb_001440_040 |
0.1878741609 |
- |
- |
hypothetical protein POPTR_0008s03520g [Populus trichocarpa] |
| 11 |
Hb_004336_040 |
0.1878766881 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 12 |
Hb_000260_560 |
0.1887726306 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g18110, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000115_160 |
0.1897673529 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g29230 [Jatropha curcas] |
| 14 |
Hb_005488_150 |
0.1906296054 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000286_170 |
0.190935786 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 16 |
Hb_048476_170 |
0.1922850239 |
- |
- |
PREDICTED: cleavage and polyadenylation specificity factor subunit 3-II-like [Citrus sinensis] |
| 17 |
Hb_002890_180 |
0.1926445295 |
- |
- |
PREDICTED: squamous cell carcinoma antigen recognized by T-cells 3 [Jatropha curcas] |
| 18 |
Hb_120470_010 |
0.1947063338 |
- |
- |
PREDICTED: PAX3- and PAX7-binding protein 1 [Jatropha curcas] |
| 19 |
Hb_000005_070 |
0.1955602319 |
transcription factor |
TF Family: PHD |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_003805_050 |
0.1956214845 |
- |
- |
- |