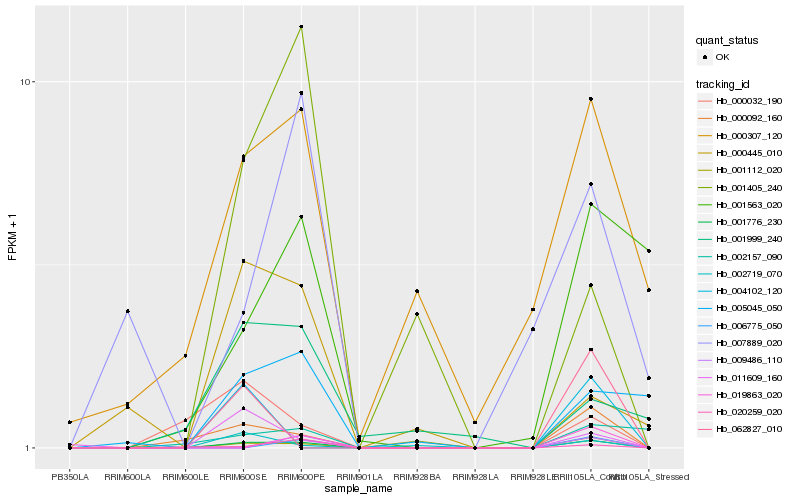
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001776_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104884260 [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_000092_160 |
0.2586166639 |
- |
- |
PREDICTED: 18 kDa seed maturation protein [Populus euphratica] |
| 3 |
Hb_011609_160 |
0.3480458168 |
- |
- |
Lipase, putative [Ricinus communis] |
| 4 |
Hb_002719_070 |
0.3550301526 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 5 |
Hb_000307_120 |
0.3607788114 |
- |
- |
hypothetical protein B456_002G120900 [Gossypium raimondii] |
| 6 |
Hb_005045_050 |
0.3635591613 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 3 [Jatropha curcas] |
| 7 |
Hb_020259_020 |
0.3681469866 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 4 [Jatropha curcas] |
| 8 |
Hb_000032_190 |
0.3887295953 |
- |
- |
- |
| 9 |
Hb_001112_020 |
0.3924029866 |
- |
- |
PREDICTED: uncharacterized protein LOC103717724, partial [Phoenix dactylifera] |
| 10 |
Hb_001405_240 |
0.3940417512 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001999_240 |
0.3977044076 |
- |
- |
PREDICTED: josephin-like protein [Populus euphratica] |
| 12 |
Hb_000445_010 |
0.3985491683 |
- |
- |
PREDICTED: uncharacterized protein LOC105113411 [Populus euphratica] |
| 13 |
Hb_001563_020 |
0.4023643238 |
transcription factor |
TF Family: TRAF |
aberrant large forked product, putative [Ricinus communis] |
| 14 |
Hb_002157_090 |
0.4043774768 |
- |
- |
hypothetical protein VITISV_041741 [Vitis vinifera] |
| 15 |
Hb_006775_050 |
0.4066877301 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g29670 isoform X2 [Nicotiana sylvestris] |
| 16 |
Hb_007889_020 |
0.4074353001 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_019863_020 |
0.4093585642 |
- |
- |
PREDICTED: wound-induced protein 1 [Jatropha curcas] |
| 18 |
Hb_004102_120 |
0.4145515126 |
- |
- |
- |
| 19 |
Hb_062827_010 |
0.4156130772 |
- |
- |
- |
| 20 |
Hb_009486_110 |
0.4161135811 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |