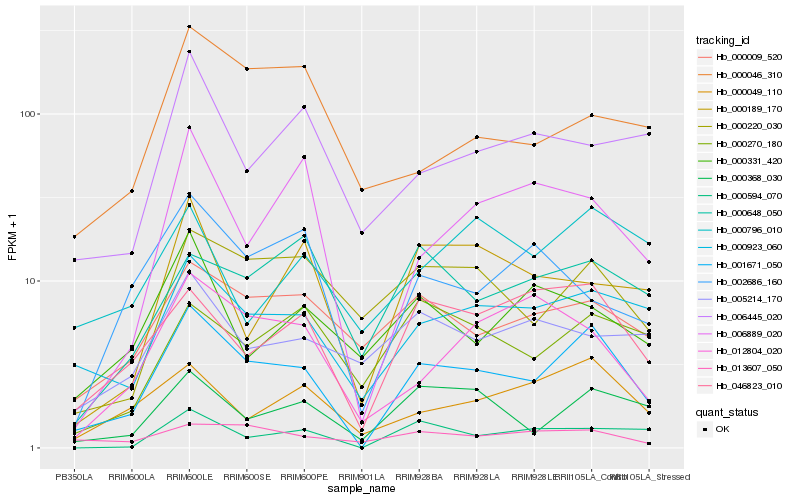
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001671_050 |
0.0 |
- |
- |
ccd1, putative [Ricinus communis] |
| 2 |
Hb_000923_060 |
0.1824166327 |
- |
- |
PREDICTED: uncharacterized protein LOC105628271 [Jatropha curcas] |
| 3 |
Hb_046823_010 |
0.1998688504 |
- |
- |
hypothetical protein CICLE_v100021301mg, partial [Citrus clementina] |
| 4 |
Hb_000368_030 |
0.2047216497 |
- |
- |
PREDICTED: cytokinin dehydrogenase 5 [Jatropha curcas] |
| 5 |
Hb_000049_110 |
0.2124599747 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.2 [Jatropha curcas] |
| 6 |
Hb_000009_520 |
0.2157213357 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 14 [Jatropha curcas] |
| 7 |
Hb_000220_030 |
0.2171534077 |
- |
- |
PREDICTED: amino-acid permease BAT1 homolog isoform X2 [Jatropha curcas] |
| 8 |
Hb_000594_070 |
0.2222476624 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_006889_020 |
0.2302588199 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 10 |
Hb_012804_020 |
0.2341425303 |
- |
- |
PREDICTED: abscisic acid 8'-hydroxylase 4-like [Jatropha curcas] |
| 11 |
Hb_013607_050 |
0.2349670711 |
- |
- |
PREDICTED: probable glycerol-3-phosphate acyltransferase 2 [Jatropha curcas] |
| 12 |
Hb_006445_020 |
0.2380040651 |
- |
- |
hypothetical protein L484_026741 [Morus notabilis] |
| 13 |
Hb_000270_180 |
0.2392241452 |
- |
- |
PREDICTED: HIPL1 protein-like [Jatropha curcas] |
| 14 |
Hb_000189_170 |
0.2413421103 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 15 |
Hb_000331_420 |
0.2443656713 |
- |
- |
PREDICTED: nucleoside diphosphate kinase 2, chloroplastic [Jatropha curcas] |
| 16 |
Hb_000648_050 |
0.2452509624 |
- |
- |
PREDICTED: putative methylesterase 14, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000796_010 |
0.2454433155 |
- |
- |
PREDICTED: nitrogen regulatory protein P-II homolog [Jatropha curcas] |
| 18 |
Hb_000046_310 |
0.2454782071 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 19 |
Hb_002686_160 |
0.2458537495 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Populus euphratica] |
| 20 |
Hb_005214_170 |
0.2460085073 |
- |
- |
PREDICTED: uncharacterized protein LOC105636021 [Jatropha curcas] |