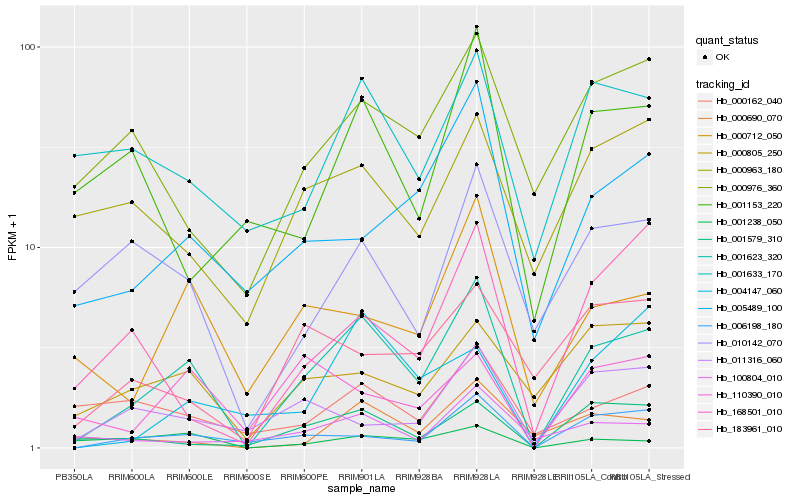
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001623_320 |
0.0 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 isoform X1 [Populus euphratica] |
| 2 |
Hb_006198_180 |
0.2326874815 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000712_050 |
0.2375579866 |
- |
- |
PREDICTED: inositol oxygenase 1-like isoform X1 [Citrus sinensis] |
| 4 |
Hb_001579_310 |
0.2420429931 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37320 [Jatropha curcas] |
| 5 |
Hb_005489_100 |
0.248191262 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000805_250 |
0.2601147902 |
- |
- |
PREDICTED: rhomboid-like protease 2 [Jatropha curcas] |
| 7 |
Hb_000690_070 |
0.2601763016 |
- |
- |
- |
| 8 |
Hb_001633_170 |
0.2616240686 |
- |
- |
PREDICTED: aspartokinase 2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001238_050 |
0.262902008 |
- |
- |
- |
| 10 |
Hb_100804_010 |
0.2634300203 |
- |
- |
BAHD family acyltransferase, clade V, putative [Theobroma cacao] |
| 11 |
Hb_010142_070 |
0.2642546765 |
- |
- |
PREDICTED: uncharacterized protein LOC105630659 [Jatropha curcas] |
| 12 |
Hb_000976_360 |
0.2653052637 |
- |
- |
PREDICTED: uncharacterized protein LOC105646237 [Jatropha curcas] |
| 13 |
Hb_168501_010 |
0.2706831711 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_110390_010 |
0.2710558622 |
- |
- |
hypothetical protein JCGZ_19741 [Jatropha curcas] |
| 15 |
Hb_000162_040 |
0.2714460158 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g21090 [Jatropha curcas] |
| 16 |
Hb_004147_060 |
0.2727653916 |
- |
- |
PREDICTED: uncharacterized protein LOC105630996 [Jatropha curcas] |
| 17 |
Hb_183961_010 |
0.2732466134 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Cucumis sativus] |
| 18 |
Hb_000963_180 |
0.2736927419 |
- |
- |
calcium/calmodulin-dependent protein kinase kinase, putative [Ricinus communis] |
| 19 |
Hb_011316_060 |
0.2743192153 |
- |
- |
alpha-l-fucosidase, putative [Ricinus communis] |
| 20 |
Hb_001153_220 |
0.2743200357 |
- |
- |
PREDICTED: UDP-galactose/UDP-glucose transporter 2 [Jatropha curcas] |