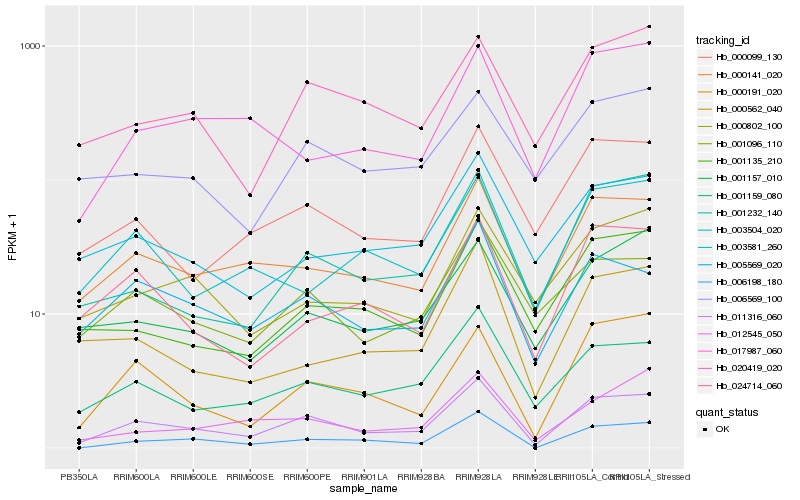
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011316_060 |
0.0 |
- |
- |
alpha-l-fucosidase, putative [Ricinus communis] |
| 2 |
Hb_000141_020 |
0.1298355316 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 6 [Pyrus x bretschneideri] |
| 3 |
Hb_001159_080 |
0.1319101016 |
- |
- |
hypothetical protein JCGZ_20745 [Jatropha curcas] |
| 4 |
Hb_006198_180 |
0.1334077663 |
- |
- |
PREDICTED: thiamine-repressible mitochondrial transport protein THI74 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005569_020 |
0.136562433 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000191_020 |
0.1453521607 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001096_110 |
0.1515549508 |
- |
- |
PREDICTED: probable VAMP-like protein At1g33475 [Jatropha curcas] |
| 8 |
Hb_001232_140 |
0.1540320496 |
- |
- |
PREDICTED: uncharacterized protein LOC105634966 [Jatropha curcas] |
| 9 |
Hb_000099_130 |
0.1549256 |
- |
- |
PREDICTED: uncharacterized protein LOC105645728 [Jatropha curcas] |
| 10 |
Hb_020419_020 |
0.1577515932 |
- |
- |
PREDICTED: calmodulin-7 isoform X1 [Vitis vinifera] |
| 11 |
Hb_024714_060 |
0.1588904089 |
- |
- |
PREDICTED: thiamine pyrophosphokinase 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_012545_050 |
0.1614931528 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Jatropha curcas] |
| 13 |
Hb_003581_260 |
0.1629186101 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 29-like [Jatropha curcas] |
| 14 |
Hb_001135_210 |
0.1635003098 |
- |
- |
PREDICTED: probable adenylate kinase 6, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001157_010 |
0.1665924215 |
- |
- |
deoxyhypusine synthase, putative [Ricinus communis] |
| 16 |
Hb_003504_020 |
0.1669424903 |
- |
- |
PREDICTED: uncharacterized protein LOC105647732 [Jatropha curcas] |
| 17 |
Hb_000562_040 |
0.1691987357 |
- |
- |
PREDICTED: auxin response factor 5 isoform X1 [Jatropha curcas] |
| 18 |
Hb_017987_060 |
0.1716212005 |
- |
- |
hypothetical protein JCGZ_15971 [Jatropha curcas] |
| 19 |
Hb_000802_100 |
0.1717956212 |
- |
- |
PREDICTED: uncharacterized protein LOC105634009 [Jatropha curcas] |
| 20 |
Hb_006569_100 |
0.1726790266 |
- |
- |
hypothetical protein POPTR_0005s26990g [Populus trichocarpa] |