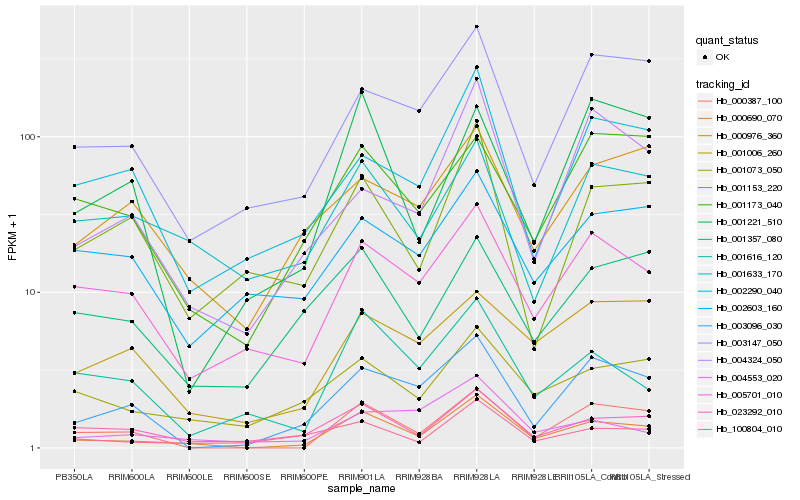
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000690_070 |
0.0 |
- |
- |
- |
| 2 |
Hb_000387_100 |
0.1649813487 |
- |
- |
- |
| 3 |
Hb_023292_010 |
0.1736700296 |
- |
- |
Cucumisin precursor, putative [Ricinus communis] |
| 4 |
Hb_003096_030 |
0.1783604259 |
- |
- |
PREDICTED: bifunctional riboflavin biosynthesis protein RIBA 1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_005701_010 |
0.1819728115 |
- |
- |
PREDICTED: uncharacterized protein LOC105633877 isoform X1 [Jatropha curcas] |
| 6 |
Hb_100804_010 |
0.1880687453 |
- |
- |
BAHD family acyltransferase, clade V, putative [Theobroma cacao] |
| 7 |
Hb_001073_050 |
0.1882183178 |
- |
- |
- |
| 8 |
Hb_003147_050 |
0.1905867163 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 9 |
Hb_001616_120 |
0.1926053752 |
- |
- |
- |
| 10 |
Hb_002290_040 |
0.2066709224 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001153_220 |
0.2067016428 |
- |
- |
PREDICTED: UDP-galactose/UDP-glucose transporter 2 [Jatropha curcas] |
| 12 |
Hb_004553_020 |
0.2088076115 |
- |
- |
PREDICTED: non-structural maintenance of chromosomes element 1 homolog [Jatropha curcas] |
| 13 |
Hb_001006_260 |
0.2098360327 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit I-like [Jatropha curcas] |
| 14 |
Hb_001357_080 |
0.2112499052 |
- |
- |
hypothetical protein POPTR_0005s15590g [Populus trichocarpa] |
| 15 |
Hb_004324_050 |
0.211889489 |
- |
- |
PREDICTED: spermine synthase [Jatropha curcas] |
| 16 |
Hb_001173_040 |
0.2145645778 |
- |
- |
PREDICTED: tryptophan--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 17 |
Hb_000976_360 |
0.215688478 |
- |
- |
PREDICTED: uncharacterized protein LOC105646237 [Jatropha curcas] |
| 18 |
Hb_001633_170 |
0.2165478595 |
- |
- |
PREDICTED: aspartokinase 2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001221_510 |
0.2198319772 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 20 |
Hb_002603_160 |
0.2216722366 |
- |
- |
Arginyl-tRNA--protein transferase, putative [Ricinus communis] |