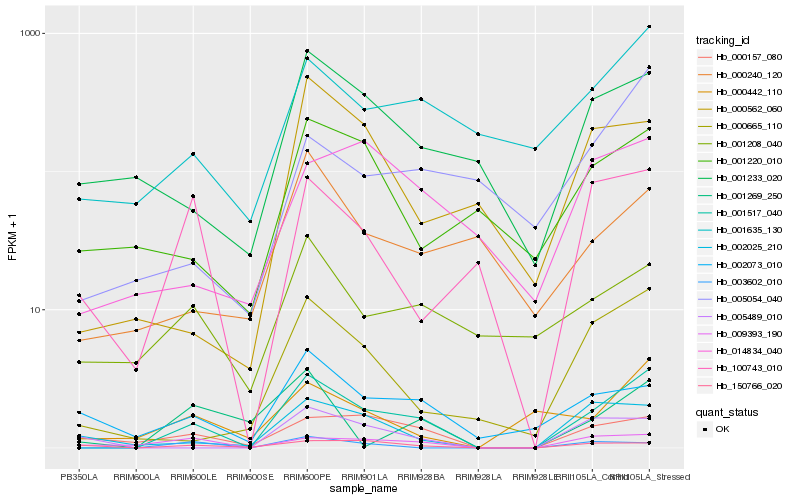
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001517_040 |
0.0 |
- |
- |
- |
| 2 |
Hb_000665_110 |
0.2599649925 |
- |
- |
Ribosomal protein S11-beta [Theobroma cacao] |
| 3 |
Hb_002025_210 |
0.2866057238 |
- |
- |
- |
| 4 |
Hb_000157_080 |
0.2869002864 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized N-acetyltransferase ycf52 [Jatropha curcas] |
| 5 |
Hb_001635_130 |
0.2915425856 |
- |
- |
PREDICTED: 60S ribosomal protein L28-2-like [Jatropha curcas] |
| 6 |
Hb_009393_190 |
0.2955194332 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase AIP2-like [Jatropha curcas] |
| 7 |
Hb_001233_020 |
0.299795716 |
- |
- |
hypothetical protein JCGZ_04109 [Jatropha curcas] |
| 8 |
Hb_005489_010 |
0.3047864462 |
- |
- |
hypothetical protein POPTR_0011s11420g [Populus trichocarpa] |
| 9 |
Hb_000562_060 |
0.3055885008 |
- |
- |
PREDICTED: uncharacterized protein LOC105169741 [Sesamum indicum] |
| 10 |
Hb_100743_010 |
0.3078686146 |
- |
- |
hypothetical protein JCGZ_17104 [Jatropha curcas] |
| 11 |
Hb_000442_110 |
0.3096554389 |
- |
- |
hypothetical protein POPTR_0010s18960g [Populus trichocarpa] |
| 12 |
Hb_002073_010 |
0.3110867046 |
- |
- |
- |
| 13 |
Hb_005054_040 |
0.3183057751 |
- |
- |
PREDICTED: 60S ribosomal protein L35a-1 [Gossypium raimondii] |
| 14 |
Hb_003602_010 |
0.318315311 |
- |
- |
serine hydroxymethyltransferase, partial [Populus alba] |
| 15 |
Hb_150766_020 |
0.3187192553 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 16 |
Hb_014834_040 |
0.318845214 |
- |
- |
hypothetical protein JCGZ_03409 [Jatropha curcas] |
| 17 |
Hb_001220_010 |
0.3245384417 |
- |
- |
hypothetical protein JCGZ_00411 [Jatropha curcas] |
| 18 |
Hb_000240_120 |
0.3250275548 |
- |
- |
ras-related protein Rab-5B [Reticulomyxa filosa] |
| 19 |
Hb_001269_250 |
0.3265007571 |
- |
- |
PREDICTED: factor of DNA methylation 1-like [Jatropha curcas] |
| 20 |
Hb_001208_040 |
0.3266352592 |
- |
- |
hypothetical protein CISIN_1g025613mg [Citrus sinensis] |