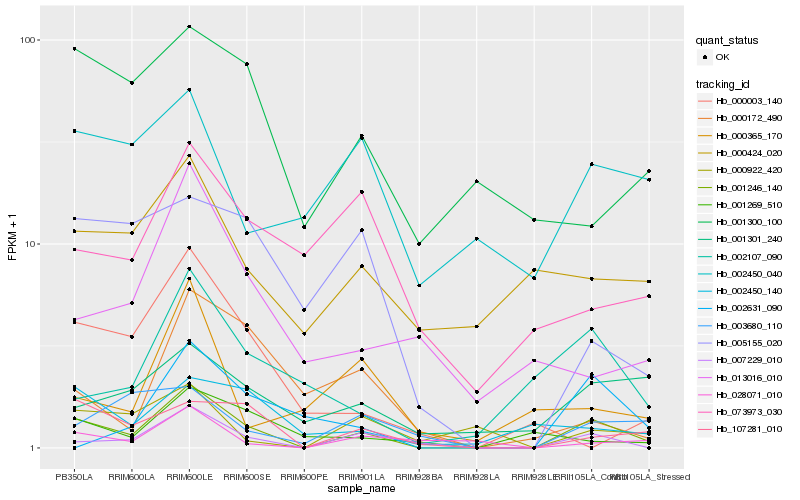
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001246_140 |
0.0 |
- |
- |
- |
| 2 |
Hb_007229_010 |
0.2541788805 |
- |
- |
PREDICTED: uncharacterized protein LOC105130855 [Populus euphratica] |
| 3 |
Hb_107281_010 |
0.2795244295 |
- |
- |
protein with unknown function [Ricinus communis] |
| 4 |
Hb_002450_140 |
0.3270164275 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 5 |
Hb_002107_090 |
0.3270744783 |
transcription factor |
TF Family: bZIP |
Basic-leucine zipper transcription factor family protein, putative [Theobroma cacao] |
| 6 |
Hb_028071_010 |
0.3273575791 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 7 |
Hb_000922_420 |
0.329657108 |
- |
- |
glutaredoxin-1, grx1, putative [Ricinus communis] |
| 8 |
Hb_001301_240 |
0.3364750626 |
- |
- |
- |
| 9 |
Hb_005155_020 |
0.3374864717 |
- |
- |
- |
| 10 |
Hb_003680_110 |
0.3388617172 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_000424_020 |
0.3458939139 |
- |
- |
Uncharacterized protein TCM_037386 [Theobroma cacao] |
| 12 |
Hb_001269_510 |
0.3475328741 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 13 |
Hb_013016_010 |
0.3515822242 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000003_140 |
0.3518772689 |
- |
- |
hypothetical protein glysoja_047034, partial [Glycine soja] |
| 15 |
Hb_001300_100 |
0.3523436602 |
- |
- |
- |
| 16 |
Hb_000172_490 |
0.3532866762 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase RHA2B [Jatropha curcas] |
| 17 |
Hb_002450_040 |
0.3547752909 |
- |
- |
PREDICTED: DNA-directed RNA polymerases II and V subunit 6B [Vitis vinifera] |
| 18 |
Hb_002631_090 |
0.3565481571 |
- |
- |
- |
| 19 |
Hb_073973_030 |
0.3578144354 |
- |
- |
PREDICTED: uncharacterized protein LOC105638905 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000365_170 |
0.3588469112 |
- |
- |
PREDICTED: 50S ribosomal protein L17, chloroplastic isoform X3 [Jatropha curcas] |