| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
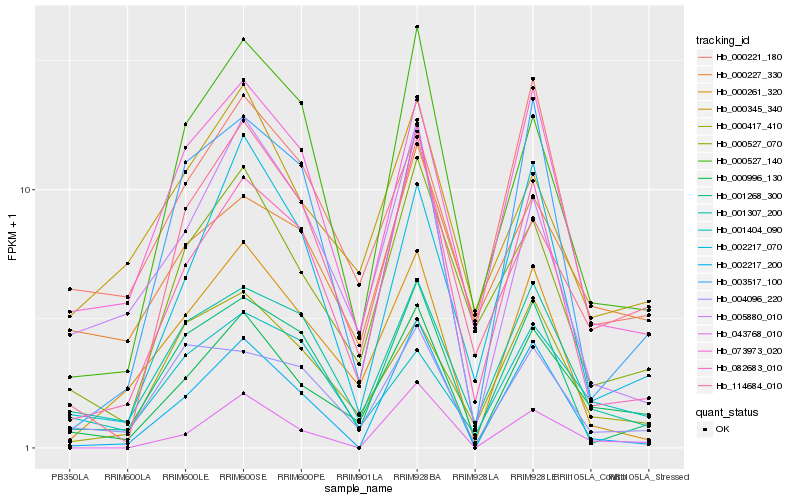
Hb_000996_130 |
0.0 |
- |
- |
PREDICTED: remorin-like [Populus euphratica] |
| 2 |
Hb_073973_020 |
0.170275748 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 3 |
Hb_001307_200 |
0.170982498 |
- |
- |
PREDICTED: uncharacterized protein LOC105644489 [Jatropha curcas] |
| 4 |
Hb_082683_010 |
0.1737084424 |
- |
- |
PREDICTED: uncharacterized protein LOC104898613 [Beta vulgaris subsp. vulgaris] |
| 5 |
Hb_000527_070 |
0.1765924681 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 6 |
Hb_114684_010 |
0.1787725314 |
- |
- |
PREDICTED: seed maturation protein PM36 [Jatropha curcas] |
| 7 |
Hb_000527_140 |
0.1814925676 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 8 |
Hb_004096_220 |
0.1817618915 |
- |
- |
PREDICTED: uncharacterized protein LOC105638706 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001268_300 |
0.1821782835 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000417_410 |
0.1821928822 |
- |
- |
PREDICTED: protein STICHEL-like 3 isoform X2 [Jatropha curcas] |
| 11 |
Hb_005880_010 |
0.1827022114 |
- |
- |
PREDICTED: uncharacterized protein LOC105646431 [Jatropha curcas] |
| 12 |
Hb_000261_320 |
0.1835107798 |
desease resistance |
Gene Name: NB-ARC |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_043768_010 |
0.1892906766 |
- |
- |
PREDICTED: heat stress transcription factor A-4a-like [Jatropha curcas] |
| 14 |
Hb_001404_090 |
0.1909948151 |
- |
- |
PREDICTED: kinesin-like protein NACK2 [Camelina sativa] |
| 15 |
Hb_002217_070 |
0.1915509125 |
- |
- |
PREDICTED: uncharacterized protein LOC105643596 [Jatropha curcas] |
| 16 |
Hb_002217_200 |
0.192951902 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 2 [Jatropha curcas] |
| 17 |
Hb_000221_180 |
0.1935926039 |
- |
- |
VQ motif-containing family protein [Populus trichocarpa] |
| 18 |
Hb_000227_330 |
0.19415222 |
- |
- |
Acyl-CoA synthetase [Ricinus communis] |
| 19 |
Hb_003517_100 |
0.1952335865 |
transcription factor |
TF Family: GRAS |
- |
| 20 |
Hb_000345_340 |
0.1956065023 |
- |
- |
PREDICTED: DNA polymerase zeta catalytic subunit [Jatropha curcas] |