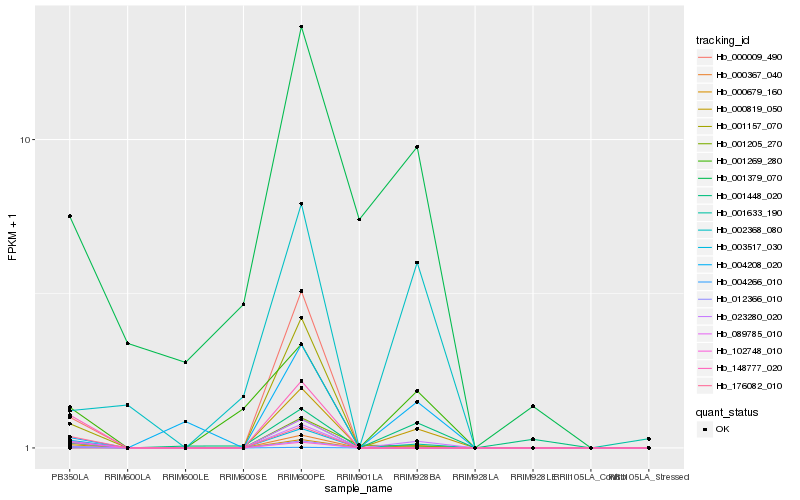
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000679_160 |
0.0 |
- |
- |
PREDICTED: probable carboxylesterase 17 [Jatropha curcas] |
| 2 |
Hb_000367_040 |
0.2371601462 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 3 |
Hb_004266_010 |
0.2590153008 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 4 |
Hb_001205_270 |
0.286466494 |
- |
- |
PREDICTED: uncharacterized protein LOC105643859 [Jatropha curcas] |
| 5 |
Hb_001269_280 |
0.2909394111 |
- |
- |
- |
| 6 |
Hb_003517_030 |
0.2974982473 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 5 isoform X2 [Eucalyptus grandis] |
| 7 |
Hb_148777_020 |
0.2983681861 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g11410 isoform X1 [Populus euphratica] |
| 8 |
Hb_176082_010 |
0.3032392186 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |
| 9 |
Hb_012366_010 |
0.3099935136 |
- |
- |
hypothetical protein JCGZ_01894 [Jatropha curcas] |
| 10 |
Hb_089785_010 |
0.3122828557 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase EFR [Fragaria vesca subsp. vesca] |
| 11 |
Hb_004208_020 |
0.3335091873 |
- |
- |
phosphoglycerate kinase, putative [Ricinus communis] |
| 12 |
Hb_001448_020 |
0.3413791332 |
- |
- |
hypothetical protein JCGZ_17204 [Jatropha curcas] |
| 13 |
Hb_001157_070 |
0.3455506485 |
- |
- |
- |
| 14 |
Hb_102748_010 |
0.345954591 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 15 |
Hb_000009_490 |
0.348067307 |
- |
- |
hypothetical protein POPTR_0007s12040g [Populus trichocarpa] |
| 16 |
Hb_001379_070 |
0.3483301595 |
- |
- |
hypothetical protein JCGZ_16377 [Jatropha curcas] |
| 17 |
Hb_002368_080 |
0.3495297952 |
- |
- |
PREDICTED: transcription factor SCREAM2-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_001633_190 |
0.3518641625 |
- |
- |
PREDICTED: probable methyltransferase PMT11 [Jatropha curcas] |
| 19 |
Hb_000819_050 |
0.3541753225 |
- |
- |
hypothetical protein Csa_4G290160 [Cucumis sativus] |
| 20 |
Hb_023280_020 |
0.3542072096 |
- |
- |
hypothetical protein JCGZ_21650 [Jatropha curcas] |