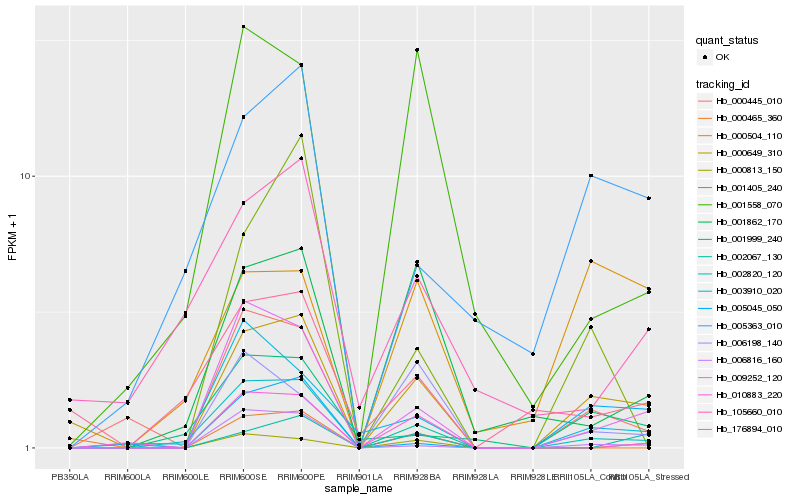
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000649_310 |
0.0 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 23 isoform X2 [Jatropha curcas] |
| 2 |
Hb_009252_120 |
0.2391200596 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002820_120 |
0.2555219263 |
transcription factor |
TF Family: ERF |
AP2 domain-containing transcription factor family protein [Populus trichocarpa] |
| 4 |
Hb_176894_010 |
0.2737840938 |
- |
- |
- |
| 5 |
Hb_006816_160 |
0.2994388912 |
- |
- |
hypothetical protein RCOM_1434920 [Ricinus communis] |
| 6 |
Hb_001999_240 |
0.301771083 |
- |
- |
PREDICTED: josephin-like protein [Populus euphratica] |
| 7 |
Hb_001862_170 |
0.3027116594 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_006198_140 |
0.3033314358 |
- |
- |
hypothetical protein L484_015208 [Morus notabilis] |
| 9 |
Hb_005045_050 |
0.3056613564 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein 3 [Jatropha curcas] |
| 10 |
Hb_000813_150 |
0.3061172305 |
- |
- |
- |
| 11 |
Hb_000465_360 |
0.3075505735 |
- |
- |
hypothetical protein CISIN_1g024854mg [Citrus sinensis] |
| 12 |
Hb_005363_010 |
0.3084084046 |
- |
- |
hypothetical protein JCGZ_13835 [Jatropha curcas] |
| 13 |
Hb_001405_240 |
0.3098609836 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003910_020 |
0.3113831604 |
- |
- |
PREDICTED: reticulon-like protein B13 [Jatropha curcas] |
| 15 |
Hb_010883_220 |
0.3171370914 |
- |
- |
PREDICTED: flavonol synthase/flavanone 3-hydroxylase-like [Prunus mume] |
| 16 |
Hb_105660_010 |
0.318969052 |
- |
- |
PREDICTED: potassium channel AKT6-like [Jatropha curcas] |
| 17 |
Hb_000445_010 |
0.3190410502 |
- |
- |
PREDICTED: uncharacterized protein LOC105113411 [Populus euphratica] |
| 18 |
Hb_001558_070 |
0.3197434417 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 19 |
Hb_000504_110 |
0.3200284037 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 20 |
Hb_002067_130 |
0.3226758127 |
- |
- |
putative non-LTR retroelement reverse transcriptase [Arabidopsis thaliana] |