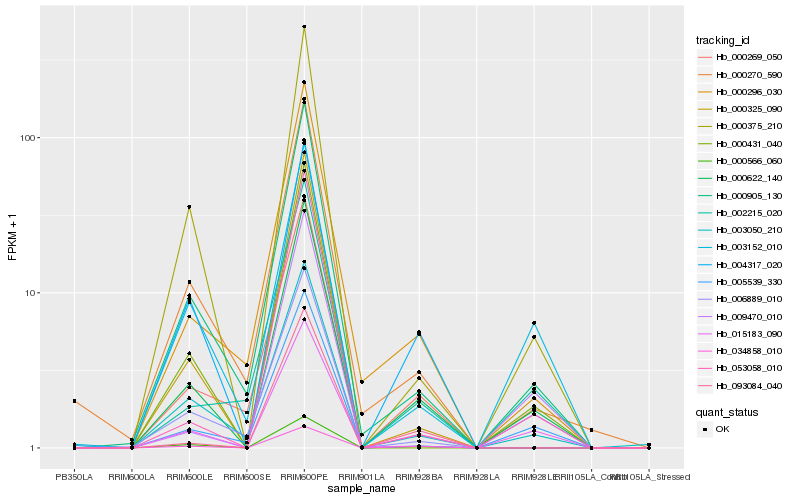
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000622_140 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000905_130 |
0.0979032624 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100 [Jatropha curcas] |
| 3 |
Hb_000375_210 |
0.1038931557 |
- |
- |
- |
| 4 |
Hb_015183_090 |
0.1088986432 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 21-like [Jatropha curcas] |
| 5 |
Hb_003050_210 |
0.113580174 |
- |
- |
PREDICTED: cytochrome b561 domain-containing protein At4g18260-like [Populus euphratica] |
| 6 |
Hb_000296_030 |
0.1140991879 |
- |
- |
PREDICTED: uncharacterized protein LOC105636578 [Jatropha curcas] |
| 7 |
Hb_000431_040 |
0.1159011811 |
- |
- |
PREDICTED: uncharacterized protein LOC105635551 [Jatropha curcas] |
| 8 |
Hb_002215_020 |
0.1176220794 |
- |
- |
hypothetical protein JCGZ_13728 [Jatropha curcas] |
| 9 |
Hb_000566_060 |
0.1200330922 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_009470_010 |
0.1224624699 |
- |
- |
- |
| 11 |
Hb_034858_010 |
0.1234813787 |
- |
- |
- |
| 12 |
Hb_006889_010 |
0.1239035712 |
- |
- |
PREDICTED: agmatine coumaroyltransferase-2-like [Jatropha curcas] |
| 13 |
Hb_004317_020 |
0.1241281776 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7-like [Malus domestica] |
| 14 |
Hb_000325_090 |
0.1249799668 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor PRE5-like [Gossypium raimondii] |
| 15 |
Hb_005539_330 |
0.1265803371 |
- |
- |
PREDICTED: WAT1-related protein At2g39510-like [Jatropha curcas] |
| 16 |
Hb_053058_010 |
0.1267229996 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_003152_010 |
0.1278826406 |
- |
- |
- |
| 18 |
Hb_000270_590 |
0.1284261387 |
- |
- |
hypothetical protein POPTR_0012s07820g [Populus trichocarpa] |
| 19 |
Hb_093084_040 |
0.1284742114 |
transcription factor |
TF Family: LOB |
hypothetical protein POPTR_0015s09370g [Populus trichocarpa] |
| 20 |
Hb_000269_050 |
0.1300080927 |
- |
- |
PREDICTED: beta-glucosidase 12-like [Gossypium raimondii] |