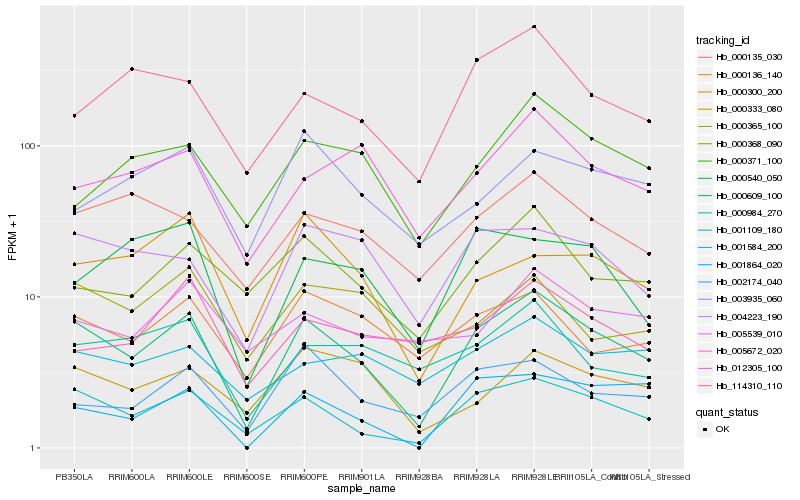
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000609_100 |
0.0 |
- |
- |
Thylakoid lumenal 16.5 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_001109_180 |
0.1011124828 |
- |
- |
PREDICTED: protein BREVIS RADIX-like [Populus euphratica] |
| 3 |
Hb_004223_190 |
0.1673968662 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 4 |
Hb_000368_090 |
0.1723010043 |
- |
- |
TPA: hypothetical protein ZEAMMB73_442906 [Zea mays] |
| 5 |
Hb_114310_110 |
0.1749284559 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 6 |
Hb_000135_030 |
0.1756260817 |
- |
- |
PREDICTED: uncharacterized protein LOC105634729 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000136_140 |
0.179045453 |
- |
- |
PREDICTED: uncharacterized protein LOC105648960 [Jatropha curcas] |
| 8 |
Hb_000333_080 |
0.1791533358 |
- |
- |
PREDICTED: uncharacterized protein LOC105637962 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002174_040 |
0.1792858413 |
- |
- |
DNAJ heat shock family protein [Theobroma cacao] |
| 10 |
Hb_000984_270 |
0.1800577749 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001864_020 |
0.1802086749 |
- |
- |
PREDICTED: transcription factor ILR3-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_005672_020 |
0.1818985586 |
- |
- |
PREDICTED: 50S ribosomal protein L6, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001584_200 |
0.1837612463 |
- |
- |
PREDICTED: Golgi apparatus membrane protein-like protein ECHIDNA [Jatropha curcas] |
| 14 |
Hb_000371_100 |
0.1839838846 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
RecName: Full=Geranylgeranyl pyrophosphate synthase, chloroplastic; Short=GGPP synthase; Short=GGPS; AltName: Full=(2E,6E)-farnesyl diphosphate synthase; AltName: Full=Dimethylallyltranstransferase; AltName: Full=Farnesyl diphosphate synthase; AltName: Full=Farnesyltranstransferase; AltName: Full=Geranyltranstransferase; Flags: Precursor [Hevea brasiliensis] |
| 15 |
Hb_000540_050 |
0.1876382664 |
- |
- |
PREDICTED: 125 kDa kinesin-related protein-like [Jatropha curcas] |
| 16 |
Hb_000300_200 |
0.1883554321 |
- |
- |
PREDICTED: xylosyltransferase 1 [Jatropha curcas] |
| 17 |
Hb_005539_010 |
0.1936621804 |
- |
- |
PREDICTED: uncharacterized protein LOC105644585 [Jatropha curcas] |
| 18 |
Hb_003935_060 |
0.1941255306 |
- |
- |
PREDICTED: very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase PASTICCINO 2 [Jatropha curcas] |
| 19 |
Hb_000365_100 |
0.1952571776 |
- |
- |
PREDICTED: probable galacturonosyltransferase 7 isoform X1 [Jatropha curcas] |
| 20 |
Hb_012305_100 |
0.1965968563 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: dihydrolipoyl dehydrogenase 2, chloroplastic-like [Jatropha curcas] |