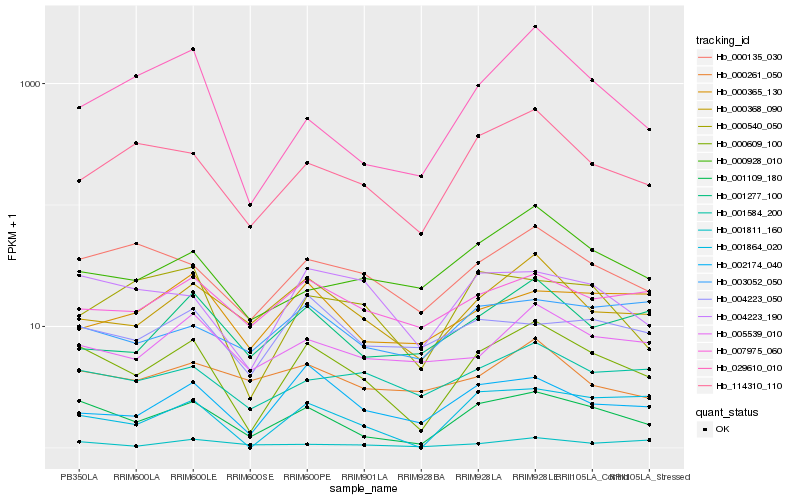
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001109_180 |
0.0 |
- |
- |
PREDICTED: protein BREVIS RADIX-like [Populus euphratica] |
| 2 |
Hb_000609_100 |
0.1011124828 |
- |
- |
Thylakoid lumenal 16.5 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_114310_110 |
0.1881089677 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 4 |
Hb_000368_090 |
0.1903823143 |
- |
- |
TPA: hypothetical protein ZEAMMB73_442906 [Zea mays] |
| 5 |
Hb_000135_030 |
0.194835326 |
- |
- |
PREDICTED: uncharacterized protein LOC105634729 isoform X1 [Jatropha curcas] |
| 6 |
Hb_004223_190 |
0.2002423649 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 7 |
Hb_003052_050 |
0.2058483757 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 8 |
Hb_001864_020 |
0.2059515524 |
- |
- |
PREDICTED: transcription factor ILR3-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_005539_010 |
0.2078387511 |
- |
- |
PREDICTED: uncharacterized protein LOC105644585 [Jatropha curcas] |
| 10 |
Hb_029610_010 |
0.2080289605 |
- |
- |
caffeic acid 3-O-methyltransferase [Hevea brasiliensis] |
| 11 |
Hb_001584_200 |
0.2082645348 |
- |
- |
PREDICTED: Golgi apparatus membrane protein-like protein ECHIDNA [Jatropha curcas] |
| 12 |
Hb_007975_060 |
0.2101293129 |
- |
- |
PREDICTED: uncharacterized GPI-anchored protein At1g61900 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001277_100 |
0.2104682867 |
- |
- |
PREDICTED: 2-methyl-6-phytyl-1,4-hydroquinone methyltransferase, chloroplastic-like isoform X1 [Gossypium raimondii] |
| 14 |
Hb_000928_010 |
0.2116428489 |
- |
- |
Cell division protein ftsZ, putative [Ricinus communis] |
| 15 |
Hb_000540_050 |
0.2117390889 |
- |
- |
PREDICTED: 125 kDa kinesin-related protein-like [Jatropha curcas] |
| 16 |
Hb_004223_050 |
0.2119900067 |
- |
- |
hexokinase [Manihot esculenta] |
| 17 |
Hb_000261_050 |
0.2120489533 |
- |
- |
PREDICTED: probable magnesium transporter NIPA2 [Populus euphratica] |
| 18 |
Hb_001811_160 |
0.2132364027 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Eucalyptus grandis] |
| 19 |
Hb_002174_040 |
0.2135424881 |
- |
- |
DNAJ heat shock family protein [Theobroma cacao] |
| 20 |
Hb_000365_130 |
0.2144113372 |
- |
- |
PREDICTED: uncharacterized protein LOC105649044 [Jatropha curcas] |