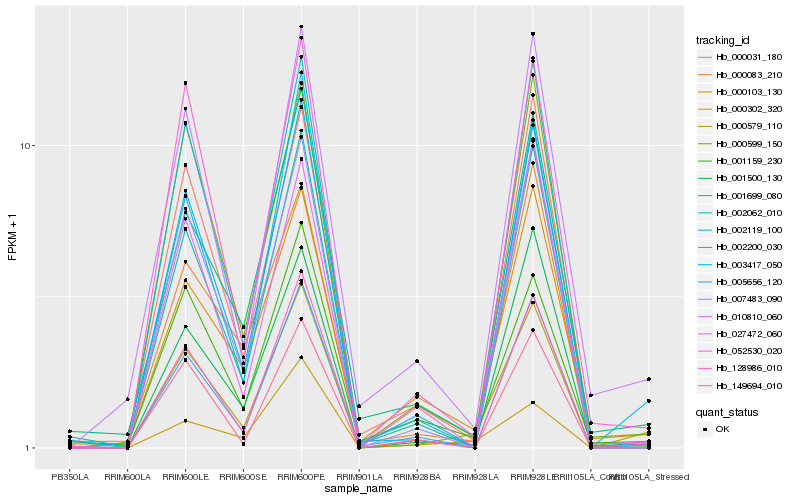
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000579_110 |
0.0 |
- |
- |
PREDICTED: transmembrane protein 136 [Jatropha curcas] |
| 2 |
Hb_002200_030 |
0.1228137523 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 3 |
Hb_003417_050 |
0.1380288547 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_005656_120 |
0.1385515138 |
- |
- |
PREDICTED: uncharacterized protein LOC105637451 [Jatropha curcas] |
| 5 |
Hb_000599_150 |
0.1405200898 |
- |
- |
PREDICTED: probable arabinose 5-phosphate isomerase [Jatropha curcas] |
| 6 |
Hb_149694_010 |
0.1431734222 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 7 |
Hb_001500_130 |
0.1449768834 |
- |
- |
interferon-induced GTP-binding protein mx, putative [Ricinus communis] |
| 8 |
Hb_010810_060 |
0.1461322432 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 9 |
Hb_007483_090 |
0.1469034402 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000103_130 |
0.1472157272 |
- |
- |
PREDICTED: protein NETWORKED 1A [Jatropha curcas] |
| 11 |
Hb_001699_080 |
0.1477172209 |
- |
- |
unnamed protein product [Coffea canephora] |
| 12 |
Hb_027472_060 |
0.1480463009 |
transcription factor |
TF Family: GRAS |
DELLA protein GAI1, putative [Ricinus communis] |
| 13 |
Hb_000302_320 |
0.1488258804 |
- |
- |
hypothetical protein JCGZ_26057 [Jatropha curcas] |
| 14 |
Hb_128986_010 |
0.1492731522 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000083_210 |
0.1506627521 |
- |
- |
Major facilitator superfamily protein, putative [Theobroma cacao] |
| 16 |
Hb_000031_180 |
0.1512933041 |
- |
- |
sarcosine oxidase, putative [Ricinus communis] |
| 17 |
Hb_052530_020 |
0.1515147455 |
- |
- |
cytochrome B5 isoform 1, putative [Ricinus communis] |
| 18 |
Hb_002119_100 |
0.1524197207 |
- |
- |
cytochrome B561, putative [Ricinus communis] |
| 19 |
Hb_002062_010 |
0.1536241926 |
- |
- |
hypothetical protein JCGZ_02762 [Jatropha curcas] |
| 20 |
Hb_001159_230 |
0.1541947916 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |