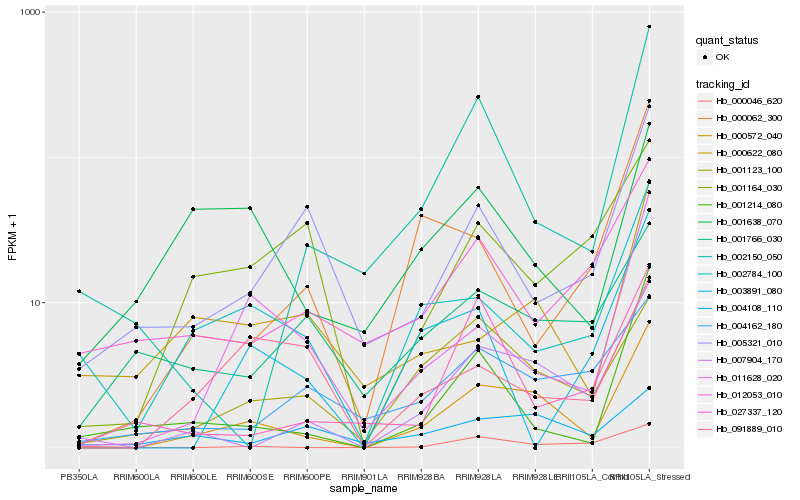
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000572_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s14815g [Populus trichocarpa] |
| 2 |
Hb_007904_170 |
0.1926304149 |
- |
- |
PREDICTED: anther-specific protein BCP1 [Jatropha curcas] |
| 3 |
Hb_001123_100 |
0.2463368771 |
- |
- |
PREDICTED: uncharacterized protein LOC105647337 isoform X1 [Jatropha curcas] |
| 4 |
Hb_011628_020 |
0.2548865482 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001214_080 |
0.2549198633 |
- |
- |
PREDICTED: transcription factor HBP-1b(c38)-like [Jatropha curcas] |
| 6 |
Hb_002784_100 |
0.2590054536 |
- |
- |
PREDICTED: uncharacterized protein LOC105631246 [Jatropha curcas] |
| 7 |
Hb_000046_620 |
0.2620816321 |
- |
- |
PREDICTED: potassium transporter 5-like [Jatropha curcas] |
| 8 |
Hb_002150_050 |
0.2725187197 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000622_080 |
0.274026821 |
- |
- |
auxin-responsive GH3 family protein [Hevea brasiliensis] |
| 10 |
Hb_004162_180 |
0.2797360259 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_001638_070 |
0.2862431293 |
transcription factor |
TF Family: bHLH |
Basic helix-loop-helix DNA-binding family protein [Theobroma cacao] |
| 12 |
Hb_004108_110 |
0.2873568972 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005321_010 |
0.2929282408 |
- |
- |
PREDICTED: uncharacterized protein LOC105644542 [Jatropha curcas] |
| 14 |
Hb_012053_010 |
0.2949262199 |
- |
- |
srpk, putative [Ricinus communis] |
| 15 |
Hb_027337_120 |
0.2950379996 |
- |
- |
PREDICTED: uncharacterized protein LOC104430249 [Eucalyptus grandis] |
| 16 |
Hb_001766_030 |
0.2958887129 |
- |
- |
hypothetical protein POPTR_0017s12640g [Populus trichocarpa] |
| 17 |
Hb_001164_030 |
0.2970236606 |
- |
- |
GMP synthase [glutamine-hydrolyzing] subunit A, putative [Ricinus communis] |
| 18 |
Hb_000062_300 |
0.2976565761 |
- |
- |
PREDICTED: uncharacterized protein LOC105644761 [Jatropha curcas] |
| 19 |
Hb_091889_010 |
0.3060849171 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 20 |
Hb_003891_080 |
0.3066213046 |
- |
- |
hypothetical protein JCGZ_16710 [Jatropha curcas] |