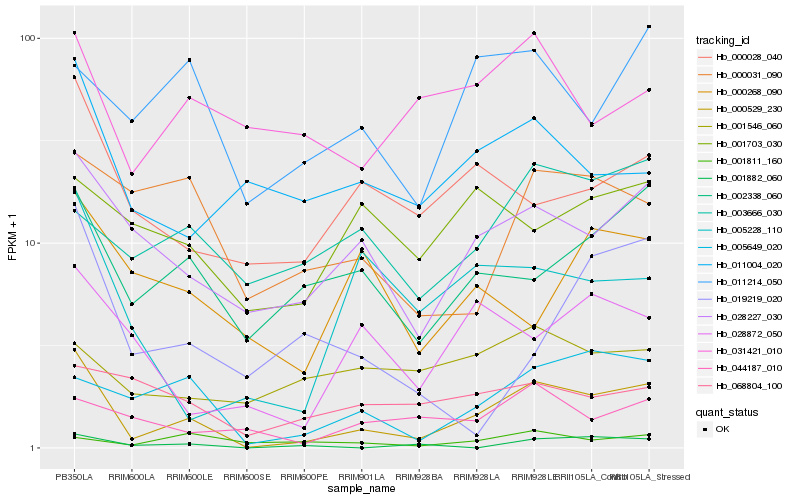
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000529_230 |
0.0 |
- |
- |
PREDICTED: putative leucine-rich repeat-containing protein DDB_G0290503 [Prunus mume] |
| 2 |
Hb_028227_030 |
0.2398179884 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005649_020 |
0.2557344399 |
- |
- |
PREDICTED: serine carboxypeptidase-like 25 [Jatropha curcas] |
| 4 |
Hb_002338_060 |
0.258890988 |
- |
- |
2Fe-2S ferredoxin [Glycine soja] |
| 5 |
Hb_000028_040 |
0.2738675272 |
- |
- |
PREDICTED: prostaglandin E synthase 2-like [Jatropha curcas] |
| 6 |
Hb_005228_110 |
0.2832474379 |
- |
- |
Cleavage and polyadenylation specificity factor subunit 3 [Fukomys damarensis] |
| 7 |
Hb_011004_020 |
0.2874191837 |
- |
- |
PREDICTED: pyridoxal 5'-phosphate synthase-like subunit PDX1.2 [Jatropha curcas] |
| 8 |
Hb_028872_050 |
0.288261177 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial [Jatropha curcas] |
| 9 |
Hb_001882_060 |
0.2919167773 |
- |
- |
- |
| 10 |
Hb_003666_030 |
0.2961250791 |
- |
- |
Thiol:disulfide interchange protein txlA, putative [Ricinus communis] |
| 11 |
Hb_011214_050 |
0.2961269289 |
- |
- |
phytoene synthase 2 [Manihot esculenta] |
| 12 |
Hb_000031_090 |
0.3001047815 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g10690 [Jatropha curcas] |
| 13 |
Hb_044187_010 |
0.3003637536 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 14 |
Hb_000268_090 |
0.3032245363 |
- |
- |
PREDICTED: uncharacterized protein LOC105628378 isoform X2 [Jatropha curcas] |
| 15 |
Hb_019219_020 |
0.3042579276 |
- |
- |
profilin-1 [Jatropha curcas] |
| 16 |
Hb_031421_010 |
0.3052142467 |
- |
- |
PREDICTED: copper chaperone for superoxide dismutase, chloroplastic/cytosolic [Jatropha curcas] |
| 17 |
Hb_068804_100 |
0.3111171748 |
- |
- |
PREDICTED: surfeit locus protein 6 [Jatropha curcas] |
| 18 |
Hb_001703_030 |
0.3137957616 |
transcription factor |
TF Family: SET |
Ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_001546_060 |
0.3143422213 |
- |
- |
PREDICTED: uncharacterized protein LOC105133943 isoform X4 [Populus euphratica] |
| 20 |
Hb_001811_160 |
0.3145558309 |
- |
- |
PREDICTED: probable glycosyltransferase At5g03795 [Eucalyptus grandis] |