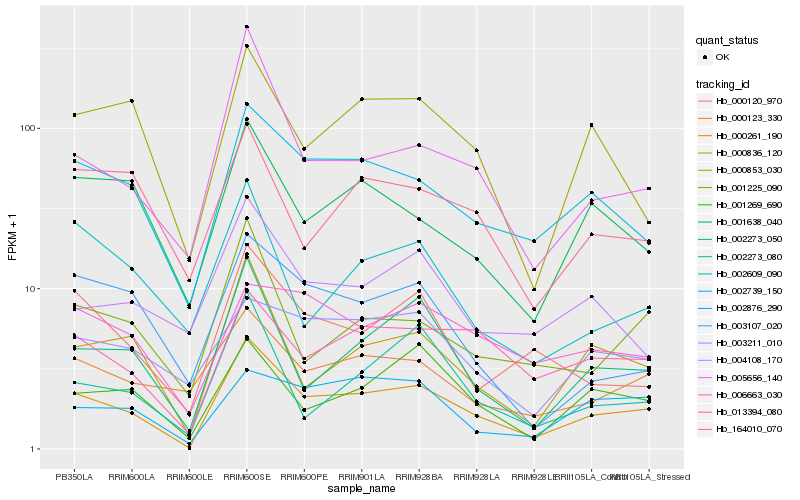
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At4g04980-like [Jatropha curcas] |
| 2 |
Hb_002739_150 |
0.1433490056 |
- |
- |
Uncharacterized protein TCM_016791 [Theobroma cacao] |
| 3 |
Hb_000836_120 |
0.1529792935 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 4 |
Hb_002273_050 |
0.1538143584 |
- |
- |
PREDICTED: geranylgeranyl diphosphate reductase, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_005656_140 |
0.1592200994 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 9 [Jatropha curcas] |
| 6 |
Hb_013394_080 |
0.1639365781 |
- |
- |
PREDICTED: putative 1-phosphatidylinositol-3-phosphate 5-kinase FAB1D isoform X2 [Jatropha curcas] |
| 7 |
Hb_000123_330 |
0.1714408829 |
- |
- |
hypothetical protein M569_14136, partial [Genlisea aurea] |
| 8 |
Hb_001225_090 |
0.1734386941 |
transcription factor |
TF Family: RWP-RK |
transcription factor, putative [Ricinus communis] |
| 9 |
Hb_001269_690 |
0.1755946059 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001638_040 |
0.1758784175 |
- |
- |
Nucleobase-ascorbate transporter 3 [Glycine soja] |
| 11 |
Hb_000853_030 |
0.1774686353 |
- |
- |
sucrose synthase 2 [Hevea brasiliensis] |
| 12 |
Hb_000120_970 |
0.1806176695 |
- |
- |
PREDICTED: probable acyl-activating enzyme 2 isoform X2 [Jatropha curcas] |
| 13 |
Hb_002273_080 |
0.1843538195 |
- |
- |
geranylgeranyl reductase [Monoraphidium neglectum] |
| 14 |
Hb_004108_170 |
0.1856579307 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_003107_020 |
0.1876014012 |
- |
- |
hypothetical protein JCGZ_18656 [Jatropha curcas] |
| 16 |
Hb_002876_290 |
0.1897256376 |
- |
- |
PREDICTED: uncharacterized protein LOC104446353 [Eucalyptus grandis] |
| 17 |
Hb_164010_070 |
0.1902030675 |
- |
- |
Paramyosin, putative [Ricinus communis] |
| 18 |
Hb_002609_090 |
0.1912481528 |
- |
- |
PREDICTED: phosphomethylpyrimidine synthase, chloroplastic isoform X2 [Jatropha curcas] |
| 19 |
Hb_003211_010 |
0.1922171808 |
- |
- |
PREDICTED: uncharacterized protein LOC105635705 [Jatropha curcas] |
| 20 |
Hb_006663_030 |
0.1924897045 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |