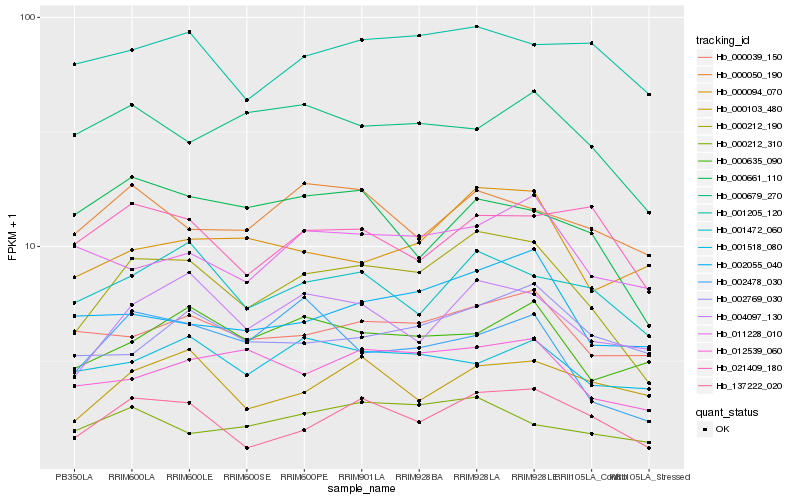
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000212_190 |
0.0 |
- |
- |
PREDICTED: phospholipase A(1) LCAT3 isoform X1 [Jatropha curcas] |
| 2 |
Hb_137222_020 |
0.1013171581 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 3 |
Hb_004097_130 |
0.1133560329 |
- |
- |
Inositol-tetrakisphosphate 1-kinase, putative [Ricinus communis] |
| 4 |
Hb_001472_060 |
0.1189387342 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8 isoform X1 [Jatropha curcas] |
| 5 |
Hb_012539_060 |
0.123300549 |
- |
- |
PREDICTED: protein EMBRYONIC FLOWER 1 [Jatropha curcas] |
| 6 |
Hb_000039_150 |
0.1275546455 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002055_040 |
0.1294719916 |
- |
- |
PREDICTED: large subunit GTPase 1 homolog [Jatropha curcas] |
| 8 |
Hb_001205_120 |
0.1331634357 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 9 |
Hb_000212_310 |
0.1333647685 |
- |
- |
hypothetical protein JCGZ_24350 [Jatropha curcas] |
| 10 |
Hb_002478_030 |
0.1338695002 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 1-like [Jatropha curcas] |
| 11 |
Hb_000094_070 |
0.1353498765 |
- |
- |
PREDICTED: 5'-nucleotidase domain-containing protein 4 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000635_090 |
0.1361435639 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 [Jatropha curcas] |
| 13 |
Hb_000679_270 |
0.1378051348 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 14 |
Hb_001518_080 |
0.1378084295 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646545 [Jatropha curcas] |
| 15 |
Hb_011228_010 |
0.1380942338 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 54, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_000661_110 |
0.1386864903 |
- |
- |
PREDICTED: protein SULFUR DEFICIENCY-INDUCED 1 [Jatropha curcas] |
| 17 |
Hb_000050_190 |
0.1387695877 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002769_030 |
0.1391515322 |
- |
- |
PREDICTED: MATE efflux family protein 4, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_021409_180 |
0.1394167374 |
- |
- |
PREDICTED: glutamine-dependent NAD(+) synthetase [Jatropha curcas] |
| 20 |
Hb_000103_480 |
0.1401940141 |
- |
- |
- |