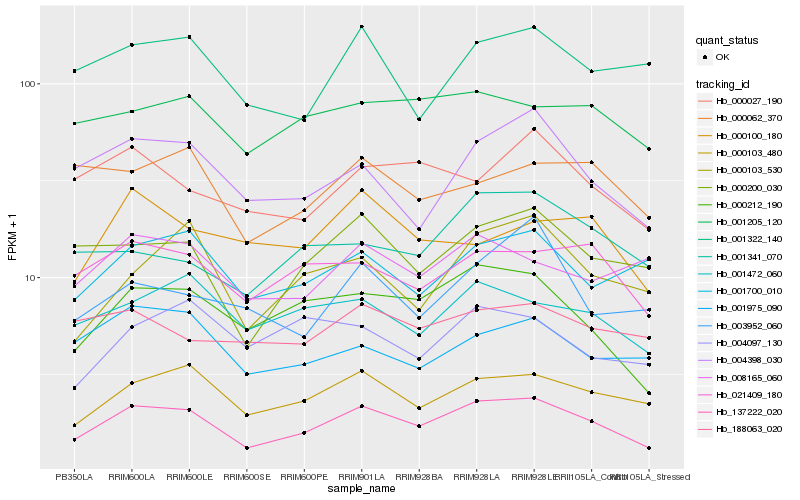
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_137222_020 |
0.0 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 2 |
Hb_000212_190 |
0.1013171581 |
- |
- |
PREDICTED: phospholipase A(1) LCAT3 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000103_480 |
0.1040008092 |
- |
- |
- |
| 4 |
Hb_000200_030 |
0.1188438467 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATC, chloroplastic [Jatropha curcas] |
| 5 |
Hb_004398_030 |
0.11906865 |
- |
- |
PREDICTED: chloroplast processing peptidase [Jatropha curcas] |
| 6 |
Hb_021409_180 |
0.1223690592 |
- |
- |
PREDICTED: glutamine-dependent NAD(+) synthetase [Jatropha curcas] |
| 7 |
Hb_000103_530 |
0.1251258508 |
- |
- |
PREDICTED: uncharacterized protein LOC105632061 [Jatropha curcas] |
| 8 |
Hb_001472_060 |
0.1253433534 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001700_010 |
0.131414216 |
- |
- |
PREDICTED: GTP-binding protein At2g22870 [Jatropha curcas] |
| 10 |
Hb_001205_120 |
0.1332592125 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] flavoprotein subunit 1, mitochondrial [Populus euphratica] |
| 11 |
Hb_000027_190 |
0.1352590538 |
- |
- |
PREDICTED: uncharacterized protein LOC105642057 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001322_140 |
0.13746644 |
- |
- |
PREDICTED: 50S ribosomal protein L19-1, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001975_090 |
0.1381606541 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor GAMYB-like [Jatropha curcas] |
| 14 |
Hb_008165_060 |
0.1403546576 |
- |
- |
PREDICTED: uncharacterized protein LOC105635541 [Jatropha curcas] |
| 15 |
Hb_000062_370 |
0.1410845823 |
- |
- |
beta-ketoacyl-ACP reductase [Vernicia fordii] |
| 16 |
Hb_001341_070 |
0.1414385094 |
- |
- |
PREDICTED: uncharacterized protein LOC105628138 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000100_180 |
0.1428230244 |
- |
- |
PREDICTED: probable acyl-activating enzyme 16, chloroplastic [Jatropha curcas] |
| 18 |
Hb_004097_130 |
0.143754172 |
- |
- |
Inositol-tetrakisphosphate 1-kinase, putative [Ricinus communis] |
| 19 |
Hb_003952_060 |
0.144441303 |
- |
- |
hypothetical protein POPTR_0016s11000g [Populus trichocarpa] |
| 20 |
Hb_188063_020 |
0.1472620995 |
- |
- |
conserved hypothetical protein [Ricinus communis] |