| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
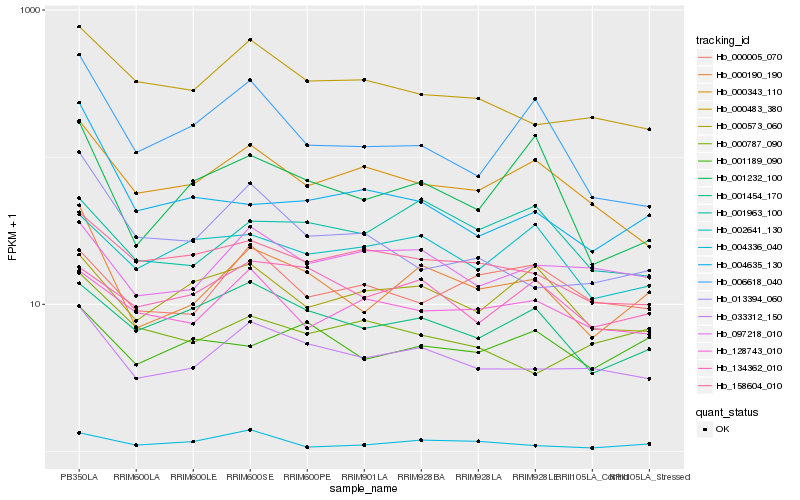
Hb_000190_190 |
0.0 |
- |
- |
PREDICTED: BI1-like protein [Gossypium raimondii] |
| 2 |
Hb_033312_150 |
0.1235313141 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 3 |
Hb_001232_100 |
0.1572369251 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 4 |
Hb_001454_170 |
0.1629680555 |
- |
- |
PREDICTED: uncharacterized protein LOC105643431 [Jatropha curcas] |
| 5 |
Hb_006618_040 |
0.1658809568 |
- |
- |
PREDICTED: hsp70-Hsp90 organizing protein 3-like [Jatropha curcas] |
| 6 |
Hb_004635_130 |
0.1668686371 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 7 |
Hb_097218_010 |
0.1670849358 |
- |
- |
PREDICTED: uncharacterized protein LOC105632664 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000343_110 |
0.1671366666 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 9 |
Hb_001189_090 |
0.1724162971 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 10 |
Hb_000483_380 |
0.1728234321 |
- |
- |
PREDICTED: bax inhibitor 1-like [Jatropha curcas] |
| 11 |
Hb_158604_010 |
0.1753244254 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 12 |
Hb_004336_040 |
0.1757272981 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 13 |
Hb_000573_060 |
0.1784794199 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2D isoform X1 [Jatropha curcas] |
| 14 |
Hb_013394_060 |
0.179069959 |
- |
- |
PREDICTED: IST1-like protein isoform X1 [Jatropha curcas] |
| 15 |
Hb_001963_100 |
0.1796438696 |
transcription factor |
TF Family: bZIP |
PREDICTED: bZIP transcription factor 17-like [Jatropha curcas] |
| 16 |
Hb_002641_130 |
0.1832948725 |
- |
- |
PREDICTED: uncharacterized protein LOC105643556 [Jatropha curcas] |
| 17 |
Hb_134362_010 |
0.1836938309 |
- |
- |
PREDICTED: FAS-associated factor 2 [Jatropha curcas] |
| 18 |
Hb_128743_010 |
0.1853587797 |
- |
- |
RNA-binding protein, putative [Ricinus communis] |
| 19 |
Hb_000005_070 |
0.1858423531 |
transcription factor |
TF Family: PHD |
protein binding protein, putative [Ricinus communis] |
| 20 |
Hb_000787_090 |
0.18660138 |
- |
- |
PREDICTED: DDRGK domain-containing protein 1 [Vitis vinifera] |