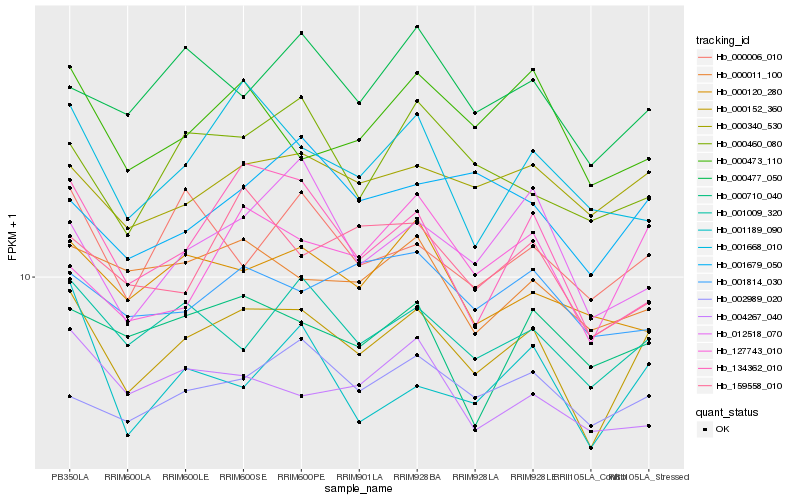
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000152_360 |
0.0 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP53 [Jatropha curcas] |
| 2 |
Hb_001009_320 |
0.0759854038 |
- |
- |
PREDICTED: uncharacterized protein LOC105640250 [Jatropha curcas] |
| 3 |
Hb_000340_530 |
0.0787761577 |
- |
- |
hypothetical protein VITISV_016664 [Vitis vinifera] |
| 4 |
Hb_134362_010 |
0.0840945142 |
- |
- |
PREDICTED: FAS-associated factor 2 [Jatropha curcas] |
| 5 |
Hb_127743_010 |
0.0845849991 |
- |
- |
PREDICTED: proteasome activator subunit 4 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002989_020 |
0.0848967426 |
- |
- |
PREDICTED: uncharacterized protein LOC105630013 isoform X2 [Jatropha curcas] |
| 7 |
Hb_000011_100 |
0.0852141052 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |
| 8 |
Hb_000477_050 |
0.0860369448 |
- |
- |
PREDICTED: 26S protease regulatory subunit 8 homolog A [Jatropha curcas] |
| 9 |
Hb_001189_090 |
0.088043525 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 10 |
Hb_000006_010 |
0.0886584673 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000473_110 |
0.0894607552 |
- |
- |
PREDICTED: PP2A regulatory subunit TAP46 [Jatropha curcas] |
| 12 |
Hb_012518_070 |
0.0900038771 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 9 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000460_080 |
0.0901713764 |
- |
- |
acyl-coenzyme A binding domain containing, putative [Ricinus communis] |
| 14 |
Hb_004267_040 |
0.0903283033 |
- |
- |
PREDICTED: transcriptional adapter ADA2a [Jatropha curcas] |
| 15 |
Hb_001814_030 |
0.0904520874 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 16 |
Hb_000710_040 |
0.0906227239 |
- |
- |
PREDICTED: HEAT repeat-containing protein 6 [Jatropha curcas] |
| 17 |
Hb_001679_050 |
0.0907530088 |
- |
- |
PREDICTED: exocyst complex component EXO84B [Jatropha curcas] |
| 18 |
Hb_159558_010 |
0.0912013565 |
- |
- |
PREDICTED: uncharacterized protein LOC105635846 [Jatropha curcas] |
| 19 |
Hb_001668_010 |
0.0913007484 |
- |
- |
PREDICTED: RNA-binding protein with multiple splicing [Jatropha curcas] |
| 20 |
Hb_000120_280 |
0.0914686443 |
- |
- |
PREDICTED: protein transport protein SEC23 [Jatropha curcas] |