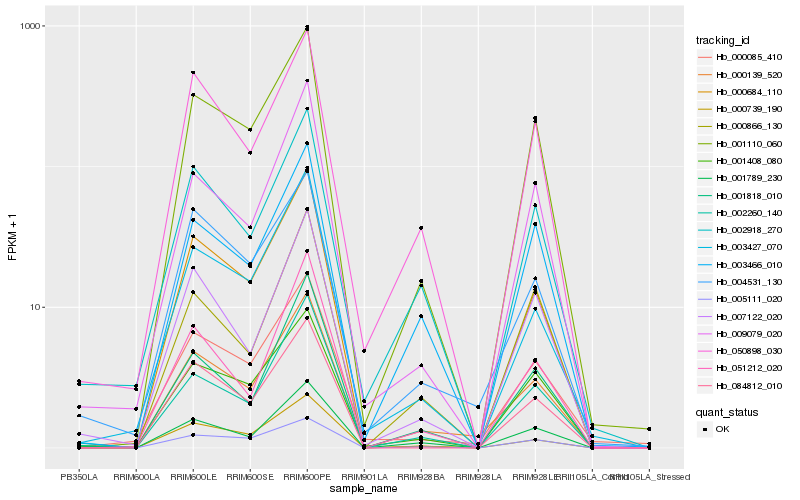
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000139_520 |
0.0 |
- |
- |
EF-hand calcium-binding domain-containing protein 4A [Theobroma cacao] |
| 2 |
Hb_001110_060 |
0.0935646173 |
- |
- |
Dehydration-responsive protein RD22 precursor, putative [Ricinus communis] |
| 3 |
Hb_000739_190 |
0.1040404182 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_003466_010 |
0.109301855 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 2.11-like [Jatropha curcas] |
| 5 |
Hb_001789_230 |
0.1143996241 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_050898_030 |
0.1146921881 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000085_410 |
0.1148947612 |
- |
- |
hypothetical protein POPTR_0011s09040g [Populus trichocarpa] |
| 8 |
Hb_002260_140 |
0.1157073432 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005111_020 |
0.116042205 |
- |
- |
hypothetical protein CISIN_1g0417821mg, partial [Citrus sinensis] |
| 10 |
Hb_001818_010 |
0.1170012074 |
- |
- |
Disease resistance response protein, putative [Ricinus communis] |
| 11 |
Hb_002918_270 |
0.1171345488 |
- |
- |
PREDICTED: protein E6-like [Populus euphratica] |
| 12 |
Hb_000684_110 |
0.1183625612 |
- |
- |
hypothetical protein POPTR_0006s13250g [Populus trichocarpa] |
| 13 |
Hb_001408_080 |
0.1184726854 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like serine/threonine-protein kinase BAM3 [Jatropha curcas] |
| 14 |
Hb_004531_130 |
0.1185381001 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA27-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_051212_020 |
0.1217035233 |
- |
- |
PREDICTED: 24-methylenesterol C-methyltransferase 2-like [Jatropha curcas] |
| 16 |
Hb_007122_020 |
0.1229139044 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 17 |
Hb_084812_010 |
0.123725098 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL21B [Vitis vinifera] |
| 18 |
Hb_003427_070 |
0.1242947995 |
- |
- |
hypothetical protein POPTR_0067s00310g [Populus trichocarpa] |
| 19 |
Hb_009079_020 |
0.1253513143 |
- |
- |
PREDICTED: protein WALLS ARE THIN 1 [Jatropha curcas] |
| 20 |
Hb_000866_130 |
0.1264830402 |
- |
- |
hypothetical protein JCGZ_14397 [Jatropha curcas] |