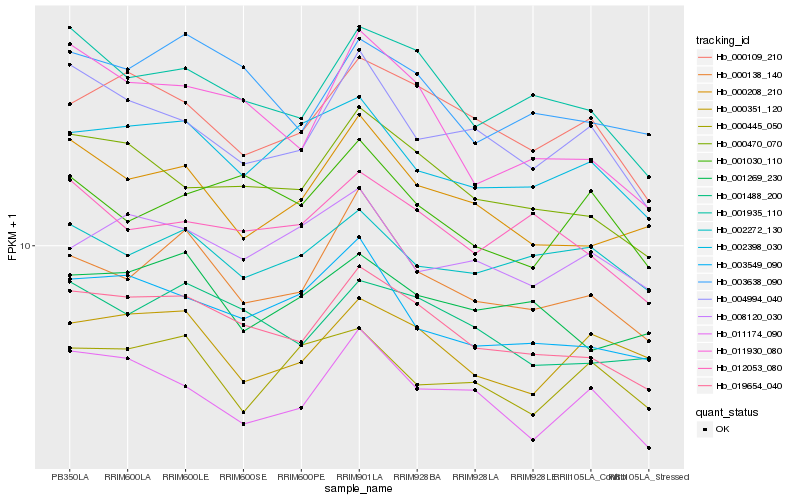
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000138_140 |
0.0 |
- |
- |
hypothetical protein B456_012G1014001, partial [Gossypium raimondii] |
| 2 |
Hb_019654_040 |
0.0761073262 |
- |
- |
PREDICTED: putative tRNA pseudouridine synthase Pus10 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000208_210 |
0.0781447086 |
- |
- |
PREDICTED: RNA-binding protein Nova-2 [Jatropha curcas] |
| 4 |
Hb_002398_030 |
0.089492321 |
- |
- |
amino acid binding protein, putative [Ricinus communis] |
| 5 |
Hb_003549_090 |
0.0910067905 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 6 [Jatropha curcas] |
| 6 |
Hb_004994_040 |
0.0936246364 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 7 |
Hb_001935_110 |
0.09448385 |
- |
- |
PREDICTED: T-complex protein 1 subunit alpha [Jatropha curcas] |
| 8 |
Hb_002272_130 |
0.09461504 |
- |
- |
PREDICTED: DNA excision repair protein ERCC-1 [Jatropha curcas] |
| 9 |
Hb_001269_230 |
0.0958111996 |
- |
- |
PREDICTED: tRNA-splicing endonuclease subunit Sen54 [Jatropha curcas] |
| 10 |
Hb_000109_210 |
0.09883773 |
- |
- |
PREDICTED: threonine--tRNA ligase, mitochondrial [Jatropha curcas] |
| 11 |
Hb_003638_090 |
0.0991546444 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 12 |
Hb_000470_070 |
0.0999140911 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP43-like [Jatropha curcas] |
| 13 |
Hb_001488_200 |
0.1005952298 |
- |
- |
PREDICTED: ribosome biogenesis protein WDR12 homolog isoform X1 [Jatropha curcas] |
| 14 |
Hb_008120_030 |
0.1016325123 |
- |
- |
PREDICTED: uncharacterized protein LOC101216675 [Cucumis sativus] |
| 15 |
Hb_000351_120 |
0.1021359003 |
- |
- |
hypothetical protein POPTR_0013s10290g [Populus trichocarpa] |
| 16 |
Hb_012053_080 |
0.1033853368 |
- |
- |
AP-2 complex subunit alpha, putative [Ricinus communis] |
| 17 |
Hb_011174_090 |
0.1037098472 |
- |
- |
PREDICTED: fidgetin-like protein 1 [Jatropha curcas] |
| 18 |
Hb_001030_110 |
0.1041928357 |
- |
- |
PREDICTED: uncharacterized PKHD-type hydroxylase At1g22950 isoform X1 [Jatropha curcas] |
| 19 |
Hb_011930_080 |
0.1049251979 |
- |
- |
PREDICTED: uncharacterized protein LOC105649790 [Jatropha curcas] |
| 20 |
Hb_000445_050 |
0.1052071706 |
- |
- |
PREDICTED: DNA topoisomerase 2-binding protein 1 [Jatropha curcas] |