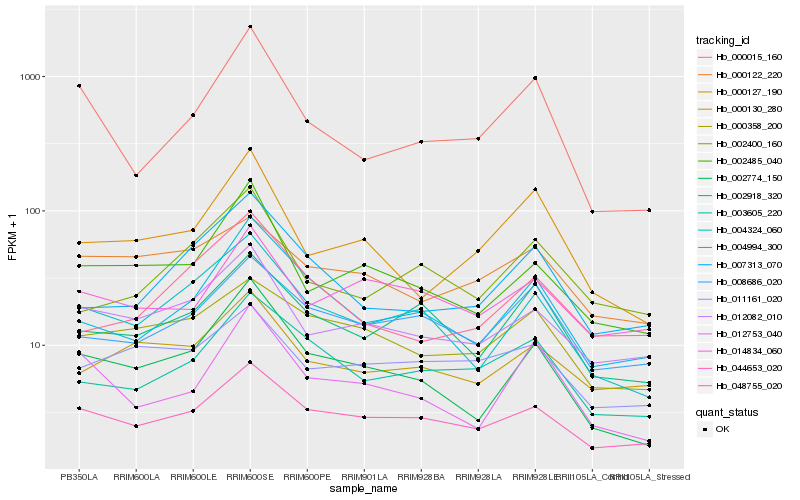
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000127_190 |
0.0 |
- |
- |
PREDICTED: protein GIGANTEA [Jatropha curcas] |
| 2 |
Hb_002485_040 |
0.1375243742 |
- |
- |
PREDICTED: uncharacterized protein LOC105637426 [Jatropha curcas] |
| 3 |
Hb_012082_010 |
0.1471826168 |
- |
- |
PREDICTED: transcription factor GTE4 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002774_150 |
0.1482847996 |
- |
- |
PREDICTED: F-box protein At1g22220 [Jatropha curcas] |
| 5 |
Hb_012753_040 |
0.1519549297 |
- |
- |
PREDICTED: F-box protein PP2-A12 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007313_070 |
0.153161125 |
- |
- |
PREDICTED: gibberellin receptor GID1C [Jatropha curcas] |
| 7 |
Hb_000015_160 |
0.1575967483 |
- |
- |
Heat shock cognate 70 kDa protein 1 [Triticum urartu] |
| 8 |
Hb_003605_220 |
0.1630305231 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 9 |
Hb_048755_020 |
0.1651673024 |
- |
- |
hypothetical protein JCGZ_09860 [Jatropha curcas] |
| 10 |
Hb_000130_280 |
0.1653093357 |
- |
- |
PREDICTED: transcription factor GTE3, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_000358_200 |
0.1655177851 |
- |
- |
PREDICTED: uncharacterized protein LOC105637771 [Jatropha curcas] |
| 12 |
Hb_008686_020 |
0.1675676682 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004994_300 |
0.1690010642 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 14 |
Hb_002400_160 |
0.1698245862 |
- |
- |
Ubiquitin-protein ligase BRE1A, putative [Ricinus communis] |
| 15 |
Hb_044653_020 |
0.1713792508 |
transcription factor, desease resistance |
TF Family: GNAT, Gene Name: NB-ARC |
hypothetical protein JCGZ_20174 [Jatropha curcas] |
| 16 |
Hb_000122_220 |
0.1722180444 |
- |
- |
Prenylated Rab acceptor protein, putative [Ricinus communis] |
| 17 |
Hb_011161_020 |
0.1737133317 |
- |
- |
PREDICTED: DIS3-like exonuclease 2 [Jatropha curcas] |
| 18 |
Hb_004324_060 |
0.1762926833 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 19 |
Hb_014834_060 |
0.1767559178 |
- |
- |
PREDICTED: uncharacterized protein LOC105632277 isoform X1 [Jatropha curcas] |
| 20 |
Hb_002918_320 |
0.1768031721 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105649589 isoform X1 [Jatropha curcas] |