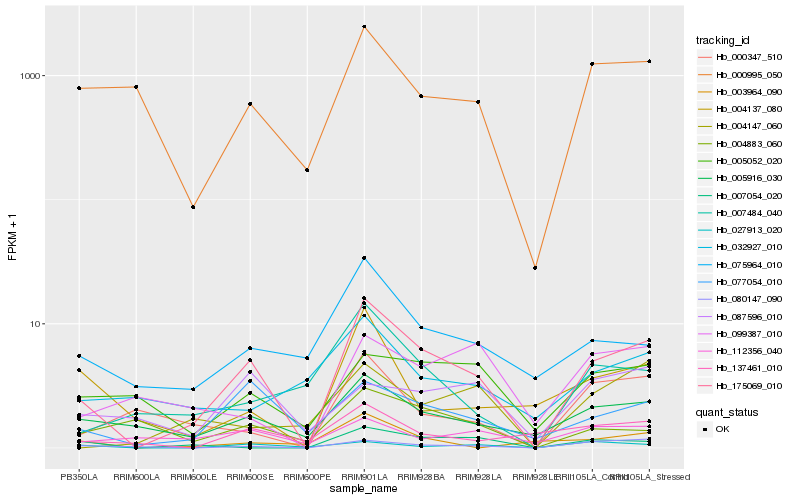
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_175069_010 |
0.0 |
- |
- |
hypothetical protein POPTR_0002s210401g, partial [Populus trichocarpa] |
| 2 |
Hb_077054_010 |
0.2488580638 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 3 |
Hb_005916_030 |
0.249859822 |
- |
- |
PREDICTED: uncharacterized protein At2g29880-like [Jatropha curcas] |
| 4 |
Hb_080147_090 |
0.271201433 |
- |
- |
hypothetical protein JCGZ_07271 [Jatropha curcas] |
| 5 |
Hb_075964_010 |
0.2781953853 |
- |
- |
hypothetical protein JCGZ_20198 [Jatropha curcas] |
| 6 |
Hb_007484_040 |
0.2847944729 |
- |
- |
- |
| 7 |
Hb_000995_050 |
0.2858673263 |
- |
- |
40S ribosomal protein S17C [Hevea brasiliensis] |
| 8 |
Hb_000347_510 |
0.2880655406 |
- |
- |
actin binding protein, putative [Ricinus communis] |
| 9 |
Hb_027913_020 |
0.2899936853 |
- |
- |
- |
| 10 |
Hb_007054_020 |
0.2936186128 |
- |
- |
- |
| 11 |
Hb_137461_010 |
0.2940513394 |
- |
- |
hypothetical protein POPTR_0011s03680g [Populus trichocarpa] |
| 12 |
Hb_005052_020 |
0.3002882411 |
- |
- |
- |
| 13 |
Hb_004883_060 |
0.3007496989 |
- |
- |
PREDICTED: uncharacterized protein LOC105647345 isoform X1 [Jatropha curcas] |
| 14 |
Hb_004137_080 |
0.3015722513 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_004147_060 |
0.3083009557 |
- |
- |
PREDICTED: uncharacterized protein LOC105630996 [Jatropha curcas] |
| 16 |
Hb_003964_090 |
0.3116944633 |
- |
- |
PREDICTED: 14-3-3-like protein GF14 iota [Jatropha curcas] |
| 17 |
Hb_099387_010 |
0.3125572884 |
- |
- |
- |
| 18 |
Hb_032927_010 |
0.3142151048 |
- |
- |
- |
| 19 |
Hb_112356_040 |
0.3154435596 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 20 |
Hb_087596_010 |
0.3170754568 |
- |
- |
PREDICTED: disease resistance protein RPS4-like [Jatropha curcas] |