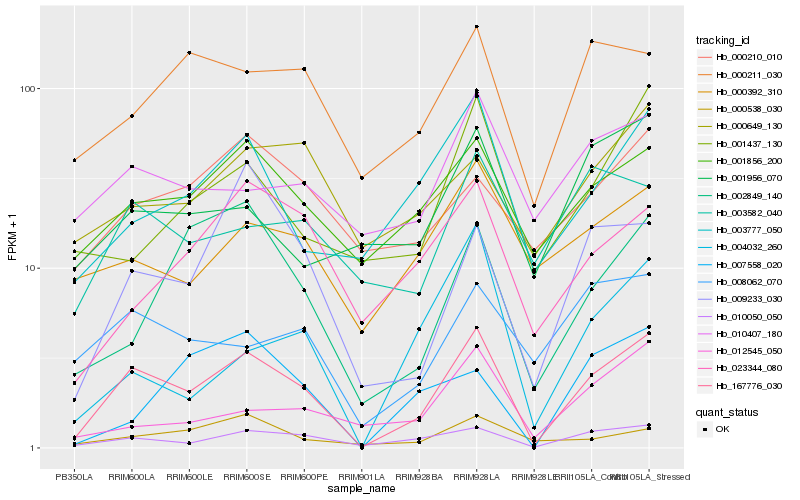
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_167776_030 |
0.0 |
- |
- |
PREDICTED: exocyst complex component EXO70A1-like [Jatropha curcas] |
| 2 |
Hb_010050_050 |
0.1719666563 |
- |
- |
PREDICTED: piezo-type mechanosensitive ion channel homolog isoform X2 [Jatropha curcas] |
| 3 |
Hb_001856_200 |
0.2215188112 |
- |
- |
Esterase PIR7B, putative [Ricinus communis] |
| 4 |
Hb_003777_050 |
0.2219788505 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105629120 [Jatropha curcas] |
| 5 |
Hb_009233_030 |
0.2250074507 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_002849_140 |
0.231872524 |
- |
- |
hypothetical protein JCGZ_14366 [Jatropha curcas] |
| 7 |
Hb_023344_080 |
0.2366807314 |
- |
- |
PREDICTED: uncharacterized protein LOC105644472 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000538_030 |
0.2389285595 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 9 |
Hb_008062_070 |
0.2389899513 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1-like [Jatropha curcas] |
| 10 |
Hb_000211_030 |
0.2396203314 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 11 |
Hb_001437_130 |
0.2422472425 |
- |
- |
hypothetical protein F383_15788 [Gossypium arboreum] |
| 12 |
Hb_000392_310 |
0.243180642 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_010407_180 |
0.2438085333 |
- |
- |
PREDICTED: phosphoenolpyruvate carboxylase kinase 1-like [Jatropha curcas] |
| 14 |
Hb_003582_040 |
0.2460089348 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0010s11160g [Populus trichocarpa] |
| 15 |
Hb_012545_050 |
0.2489902822 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Jatropha curcas] |
| 16 |
Hb_001956_070 |
0.2509826508 |
- |
- |
PREDICTED: telomere repeat-binding protein 2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000210_010 |
0.2512237821 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 11 [Jatropha curcas] |
| 18 |
Hb_007558_020 |
0.2532306345 |
- |
- |
hypothetical protein JCGZ_17631 [Jatropha curcas] |
| 19 |
Hb_000649_130 |
0.2538005857 |
- |
- |
Polcalcin Jun o, putative [Ricinus communis] |
| 20 |
Hb_004032_260 |
0.2555333963 |
- |
- |
PREDICTED: uncharacterized protein LOC105635147 [Jatropha curcas] |