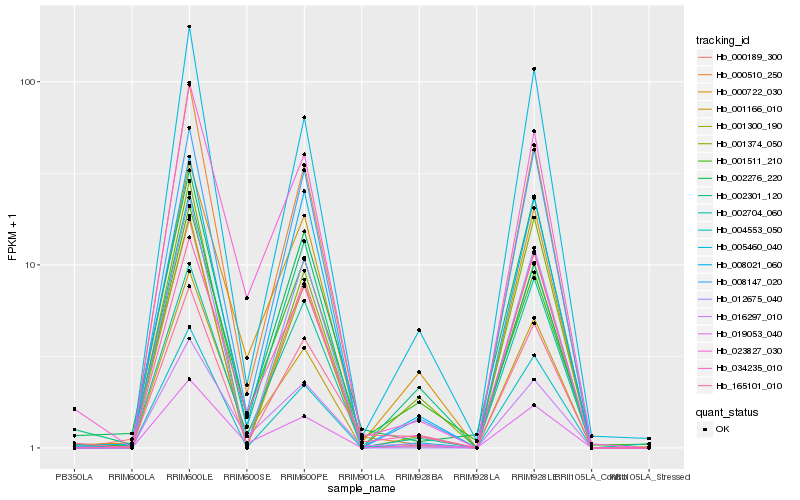
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_165101_010 |
0.0 |
- |
- |
PREDICTED: putative wall-associated receptor kinase-like 16 [Nelumbo nucifera] |
| 2 |
Hb_016297_010 |
0.1002883432 |
- |
- |
PREDICTED: uncharacterized protein LOC105638665 [Jatropha curcas] |
| 3 |
Hb_002276_220 |
0.1012951049 |
- |
- |
PREDICTED: uncharacterized protein LOC105640684 isoform X2 [Jatropha curcas] |
| 4 |
Hb_005460_040 |
0.1079602508 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 2 [Jatropha curcas] |
| 5 |
Hb_001300_190 |
0.1111398743 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_004553_050 |
0.1114107871 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g10020 [Jatropha curcas] |
| 7 |
Hb_008021_060 |
0.1116161756 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 8 |
Hb_001511_210 |
0.1138700785 |
- |
- |
PREDICTED: protein ESKIMO 1-like [Jatropha curcas] |
| 9 |
Hb_008147_020 |
0.1165091063 |
- |
- |
hypothetical protein JCGZ_25652 [Jatropha curcas] |
| 10 |
Hb_034235_010 |
0.1169367322 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 11 |
Hb_000510_250 |
0.118409039 |
transcription factor |
TF Family: AUX/IAA |
auxin-induced protein aux28 [Populus trichocarpa] |
| 12 |
Hb_002704_060 |
0.1222050527 |
- |
- |
PREDICTED: probable inactive leucine-rich repeat receptor-like protein kinase At1g66830 [Jatropha curcas] |
| 13 |
Hb_000722_030 |
0.1225543324 |
- |
- |
putative plant disease resistance family protein [Populus trichocarpa] |
| 14 |
Hb_023827_030 |
0.12273322 |
- |
- |
PREDICTED: thylakoid lumenal 16.5 kDa protein, chloroplastic-like [Populus euphratica] |
| 15 |
Hb_019053_040 |
0.1233044066 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_000189_300 |
0.1238893592 |
transcription factor |
TF Family: Trihelix |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_001374_050 |
0.125355397 |
- |
- |
PREDICTED: subtilisin-like protease SBT5.3 [Jatropha curcas] |
| 18 |
Hb_012675_040 |
0.1261455383 |
- |
- |
PREDICTED: uncharacterized protein LOC105647773 isoform X1 [Jatropha curcas] |
| 19 |
Hb_002301_120 |
0.1264826819 |
- |
- |
PREDICTED: uncharacterized protein LOC105649581 [Jatropha curcas] |
| 20 |
Hb_001166_010 |
0.1281125786 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |