| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
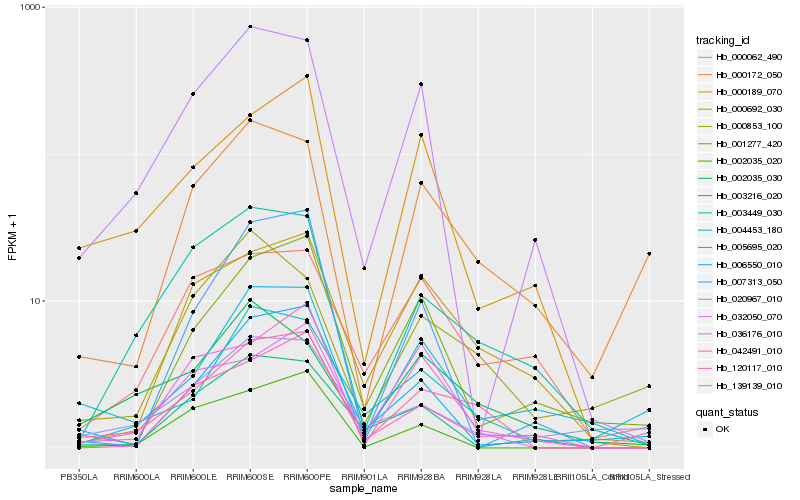
Hb_139139_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003449_030 |
0.1878596335 |
- |
- |
PREDICTED: 5'-adenylylsulfate reductase 3, chloroplastic-like [Camelina sativa] |
| 3 |
Hb_000692_030 |
0.1895155969 |
- |
- |
nucleoredoxin, putative [Ricinus communis] |
| 4 |
Hb_042491_010 |
0.1939836095 |
- |
- |
JHL06P13.14 [Jatropha curcas] |
| 5 |
Hb_000189_070 |
0.1995264238 |
transcription factor |
TF Family: AUX/IAA |
Auxin-induced protein 22D, putative [Ricinus communis] |
| 6 |
Hb_005695_020 |
0.2058156454 |
- |
- |
PREDICTED: low affinity sulfate transporter 3 [Jatropha curcas] |
| 7 |
Hb_002035_020 |
0.2170986913 |
transcription factor |
TF Family: TRAF |
hypothetical protein B456_011G050200 [Gossypium raimondii] |
| 8 |
Hb_032050_070 |
0.2199043076 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 9 |
Hb_006550_010 |
0.2200642886 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Eucalyptus grandis] |
| 10 |
Hb_036176_010 |
0.2216313871 |
- |
- |
hypothetical protein CISIN_1g002055mg [Citrus sinensis] |
| 11 |
Hb_002035_030 |
0.2255730089 |
- |
- |
hypothetical protein PRUPE_ppa003221mg [Prunus persica] |
| 12 |
Hb_000172_050 |
0.2267844269 |
- |
- |
PREDICTED: uncharacterized protein LOC105650611 [Jatropha curcas] |
| 13 |
Hb_001277_420 |
0.2278443328 |
- |
- |
PREDICTED: protein lin-12-like isoform X2 [Populus euphratica] |
| 14 |
Hb_020967_010 |
0.2302349441 |
- |
- |
- |
| 15 |
Hb_000062_490 |
0.2304066912 |
- |
- |
PREDICTED: lipoxygenase 6, chloroplastic [Jatropha curcas] |
| 16 |
Hb_120117_010 |
0.230610192 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 17 |
Hb_003216_020 |
0.2317977869 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 2 [Jatropha curcas] |
| 18 |
Hb_004453_180 |
0.2329826853 |
- |
- |
hypothetical protein JCGZ_07188 [Jatropha curcas] |
| 19 |
Hb_000853_100 |
0.2355095159 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF073 [Jatropha curcas] |
| 20 |
Hb_007313_050 |
0.2361242702 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |