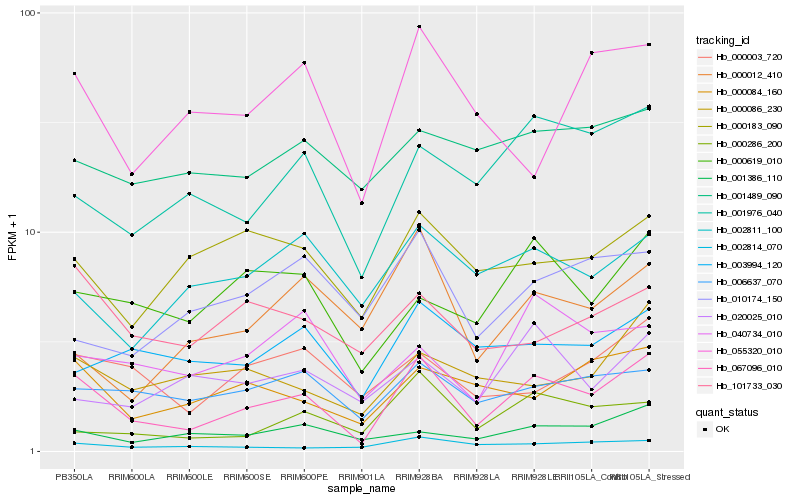
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_067096_010 |
0.0 |
- |
- |
PREDICTED: valine--tRNA ligase [Jatropha curcas] |
| 2 |
Hb_001976_040 |
0.1870322459 |
- |
- |
PREDICTED: nicotinamidase 1-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_006637_070 |
0.1953480369 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 4 |
Hb_000084_160 |
0.1974891046 |
- |
- |
SGNH hydrolase-type esterase superfamily protein, putative [Theobroma cacao] |
| 5 |
Hb_000619_010 |
0.199981644 |
- |
- |
TPA: hypothetical protein ZEAMMB73_187549 [Zea mays] |
| 6 |
Hb_000286_200 |
0.2005429983 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X2 [Jatropha curcas] |
| 7 |
Hb_002814_070 |
0.2010063466 |
- |
- |
PREDICTED: uncharacterized protein LOC104877459 [Vitis vinifera] |
| 8 |
Hb_001386_110 |
0.2041171612 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: putative fasciclin-like arabinogalactan protein 20 [Populus euphratica] |
| 9 |
Hb_000003_720 |
0.2051873595 |
- |
- |
hypothetical protein POPTR_0003s04720g [Populus trichocarpa] |
| 10 |
Hb_000086_230 |
0.2063053523 |
- |
- |
serine/threonine-protein kinase, putative [Ricinus communis] |
| 11 |
Hb_003994_120 |
0.2097153147 |
- |
- |
PREDICTED: uncharacterized protein LOC105130433 isoform X1 [Populus euphratica] |
| 12 |
Hb_000183_090 |
0.2121655894 |
- |
- |
PREDICTED: protein EARLY FLOWERING 3 isoform X2 [Jatropha curcas] |
| 13 |
Hb_101733_030 |
0.2134532608 |
- |
- |
PREDICTED: uncharacterized protein At3g49055 [Jatropha curcas] |
| 14 |
Hb_055320_010 |
0.213477671 |
- |
- |
PREDICTED: oligosaccharyltransferase complex subunit ostc [Jatropha curcas] |
| 15 |
Hb_040734_010 |
0.2137533337 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37230 [Jatropha curcas] |
| 16 |
Hb_002811_100 |
0.2184749556 |
- |
- |
PREDICTED: uncharacterized protein LOC105647658 [Jatropha curcas] |
| 17 |
Hb_000012_410 |
0.2193987846 |
- |
- |
PREDICTED: histone deacetylase 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_020025_010 |
0.2220794464 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Jatropha curcas] |
| 19 |
Hb_001489_090 |
0.2225243052 |
- |
- |
PREDICTED: uncharacterized protein LOC105649775 [Jatropha curcas] |
| 20 |
Hb_010174_150 |
0.2250359641 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |