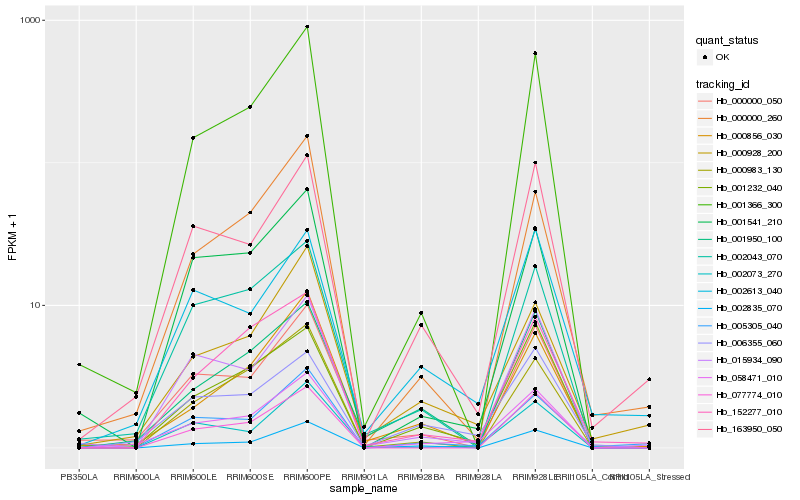
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_058471_010 |
0.0 |
- |
- |
PREDICTED: clathrin light chain 1 [Jatropha curcas] |
| 2 |
Hb_006355_060 |
0.1322081909 |
- |
- |
PREDICTED: transcription factor PIF1-like isoform X2 [Jatropha curcas] |
| 3 |
Hb_001366_300 |
0.1332150667 |
- |
- |
GERMIN-LIKE protein 10 [Populus trichocarpa] |
| 4 |
Hb_000000_050 |
0.137175824 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001232_040 |
0.1372670922 |
- |
- |
PREDICTED: uncharacterized protein LOC105634974 [Jatropha curcas] |
| 6 |
Hb_005305_040 |
0.1373581667 |
transcription factor |
TF Family: C3H |
transcription factor, putative [Ricinus communis] |
| 7 |
Hb_163950_050 |
0.1375840744 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 8 |
Hb_015934_090 |
0.1409486525 |
- |
- |
PREDICTED: uncharacterized protein LOC105640158 [Jatropha curcas] |
| 9 |
Hb_002073_270 |
0.1442283704 |
desease resistance |
Gene Name: TIR |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 10 |
Hb_001950_100 |
0.1460523828 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g37250 [Jatropha curcas] |
| 11 |
Hb_002043_070 |
0.1475111253 |
- |
- |
PREDICTED: jacalin-related lectin 19 [Jatropha curcas] |
| 12 |
Hb_002835_070 |
0.1519017632 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000928_200 |
0.1523828868 |
- |
- |
PREDICTED: RING-H2 finger protein ATL52-like [Jatropha curcas] |
| 14 |
Hb_001541_210 |
0.1533402804 |
- |
- |
hypothetical protein POPTR_0008s19020g [Populus trichocarpa] |
| 15 |
Hb_000000_260 |
0.1552705471 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 1 [Jatropha curcas] |
| 16 |
Hb_002613_040 |
0.1570033569 |
- |
- |
phosphate translocator-related family protein [Populus trichocarpa] |
| 17 |
Hb_000983_130 |
0.1570450698 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 18 |
Hb_152277_010 |
0.1571259231 |
- |
- |
PREDICTED: 9-cis-epoxycarotenoid dioxygenase NCED3, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_077774_010 |
0.1578704147 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 20 |
Hb_000856_030 |
0.1580322253 |
- |
- |
ATP binding protein, putative [Ricinus communis] |