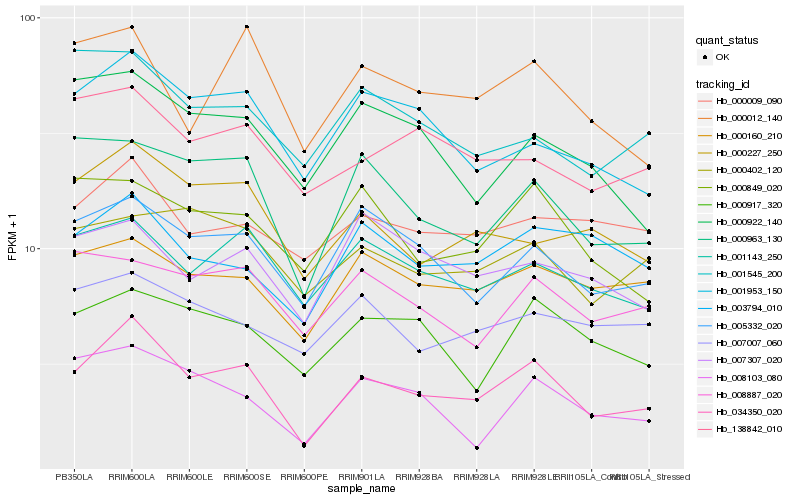
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_034350_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636191 [Jatropha curcas] |
| 2 |
Hb_000963_130 |
0.1166887164 |
- |
- |
RNA m5u methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_005332_020 |
0.1201396903 |
transcription factor |
TF Family: SNF2 |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 4 |
Hb_001953_150 |
0.1236187775 |
- |
- |
PREDICTED: protein DEK isoform X2 [Jatropha curcas] |
| 5 |
Hb_000227_250 |
0.1260883245 |
transcription factor |
TF Family: SNF2 |
PREDICTED: probable ATP-dependent DNA helicase CHR12 [Jatropha curcas] |
| 6 |
Hb_001143_250 |
0.1271973157 |
- |
- |
PREDICTED: SWI/SNF complex subunit SWI3C isoform X1 [Jatropha curcas] |
| 7 |
Hb_000012_140 |
0.1312260659 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_008103_080 |
0.1316855762 |
- |
- |
PREDICTED: lycopene epsilon cyclase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000160_210 |
0.1322810785 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000917_320 |
0.1344514105 |
- |
- |
PREDICTED: uncharacterized protein LOC105641975 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003794_010 |
0.1362658538 |
- |
- |
PREDICTED: uncharacterized protein LOC105641013 [Jatropha curcas] |
| 12 |
Hb_000849_020 |
0.1363606477 |
- |
- |
PREDICTED: uncharacterized protein LOC105644228 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000009_090 |
0.1369220852 |
transcription factor |
TF Family: bZIP |
G-box-binding factor, putative [Ricinus communis] |
| 14 |
Hb_138842_010 |
0.1377742309 |
transcription factor |
TF Family: Orphans |
PREDICTED: ethylene receptor isoform X2 [Jatropha curcas] |
| 15 |
Hb_000402_120 |
0.1405270688 |
- |
- |
PREDICTED: uncharacterized protein LOC105634656 [Jatropha curcas] |
| 16 |
Hb_008887_020 |
0.1414328397 |
- |
- |
PREDICTED: uncharacterized protein LOC105636191 [Jatropha curcas] |
| 17 |
Hb_000922_140 |
0.1414619253 |
- |
- |
PREDICTED: peroxisomal membrane protein PEX14 [Jatropha curcas] |
| 18 |
Hb_001545_200 |
0.1445358144 |
- |
- |
PREDICTED: uncharacterized oxidoreductase At4g09670 [Jatropha curcas] |
| 19 |
Hb_007007_060 |
0.144581352 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_007307_020 |
0.1459218518 |
- |
- |
DNA binding protein, putative [Ricinus communis] |