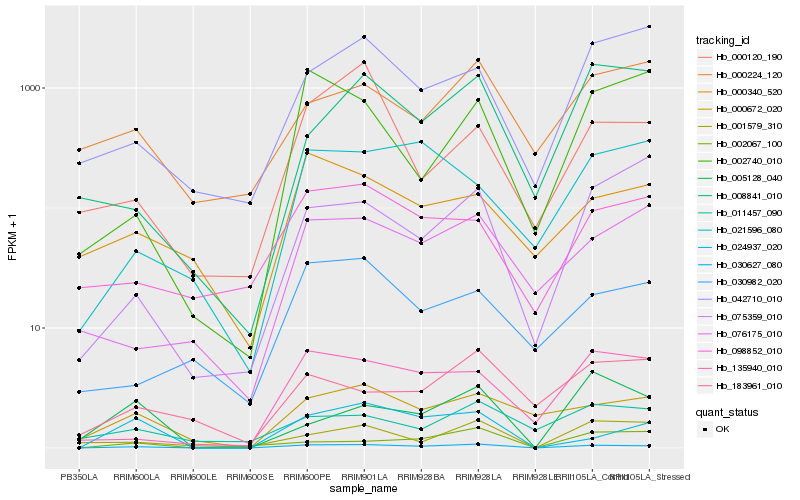
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030627_080 |
0.0 |
- |
- |
protein with unknown function [Ricinus communis] |
| 2 |
Hb_024937_020 |
0.2109592481 |
- |
- |
- |
| 3 |
Hb_135940_010 |
0.2313857192 |
- |
- |
hypothetical protein JCGZ_18294 [Jatropha curcas] |
| 4 |
Hb_000672_020 |
0.2431717216 |
- |
- |
Peptidoglycan-binding LysM domain-containing protein, putative isoform 3 [Theobroma cacao] |
| 5 |
Hb_002067_100 |
0.2454747439 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001579_310 |
0.2463802296 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g37320 [Jatropha curcas] |
| 7 |
Hb_075359_010 |
0.2528020666 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 8 |
Hb_076175_010 |
0.2537615451 |
- |
- |
PREDICTED: DNA-directed RNA polymerase II subunit RPB7 [Cucumis melo] |
| 9 |
Hb_005128_040 |
0.2556458303 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002740_010 |
0.2557476478 |
- |
- |
stress-induced hydrophobic peptide [Cleistogenes songorica] |
| 11 |
Hb_042710_010 |
0.2600976505 |
- |
- |
PREDICTED: 40S ribosomal protein S15a-1 [Jatropha curcas] |
| 12 |
Hb_030982_020 |
0.2646281625 |
- |
- |
PREDICTED: ubiquitin domain-containing protein 1 [Jatropha curcas] |
| 13 |
Hb_098852_010 |
0.2649811662 |
- |
- |
PREDICTED: MATH domain-containing protein At5g43560-like [Jatropha curcas] |
| 14 |
Hb_008841_010 |
0.2652187241 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |
| 15 |
Hb_021596_080 |
0.2680308722 |
- |
- |
50S ribosomal protein L33A [Hevea brasiliensis] |
| 16 |
Hb_183961_010 |
0.2695693022 |
- |
- |
PREDICTED: inactive rhomboid protein 1 [Cucumis sativus] |
| 17 |
Hb_000340_520 |
0.2706606563 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 5C [Jatropha curcas] |
| 18 |
Hb_000224_120 |
0.270978213 |
- |
- |
eukaryotic translation initiation factor 1Aa [Hevea brasiliensis] |
| 19 |
Hb_011457_090 |
0.2732690039 |
- |
- |
PREDICTED: methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial [Jatropha curcas] |
| 20 |
Hb_000120_190 |
0.2742303464 |
- |
- |
Pre-mRNA-splicing factor ini1 [Ricinus communis] |