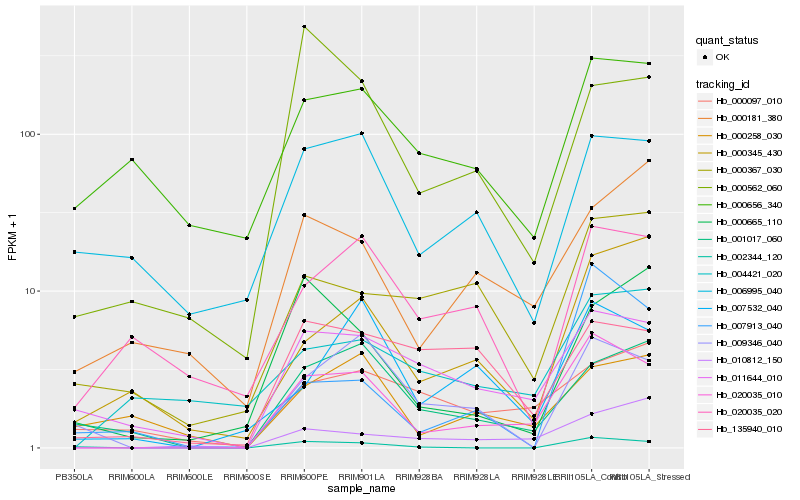
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_020035_010 |
0.0 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 1-like isoform X2 [Glycine max] |
| 2 |
Hb_011644_010 |
0.2312424796 |
- |
- |
PREDICTED: probable choline-phosphate cytidylyltransferase [Elaeis guineensis] |
| 3 |
Hb_002344_120 |
0.2358832286 |
- |
- |
- |
| 4 |
Hb_009346_040 |
0.244925791 |
- |
- |
- |
| 5 |
Hb_007532_040 |
0.2462584497 |
- |
- |
- |
| 6 |
Hb_010812_150 |
0.2482784168 |
- |
- |
hypothetical protein CICLE_v10009763mg [Citrus clementina] |
| 7 |
Hb_000345_430 |
0.257823657 |
- |
- |
40S ribosomal protein S24A [Hevea brasiliensis] |
| 8 |
Hb_001017_060 |
0.2591191427 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 9 |
Hb_000367_030 |
0.271666304 |
- |
- |
hypothetical protein POPTR_0015s06690g [Populus trichocarpa] |
| 10 |
Hb_004421_020 |
0.2741001938 |
- |
- |
- |
| 11 |
Hb_007913_040 |
0.2787112115 |
- |
- |
sterol desaturase, putative [Ricinus communis] |
| 12 |
Hb_020035_020 |
0.2788123395 |
- |
- |
PREDICTED: uncharacterized protein LOC105631020 [Jatropha curcas] |
| 13 |
Hb_006995_040 |
0.2805070796 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 14 kDa zinc-binding protein-like [Pyrus x bretschneideri] |
| 14 |
Hb_000181_380 |
0.2805104655 |
- |
- |
PREDICTED: cytochrome c oxidase copper chaperone 1 [Jatropha curcas] |
| 15 |
Hb_135940_010 |
0.2812642651 |
- |
- |
hypothetical protein JCGZ_18294 [Jatropha curcas] |
| 16 |
Hb_000258_030 |
0.2839101428 |
- |
- |
- |
| 17 |
Hb_000656_340 |
0.2843856224 |
- |
- |
hypothetical protein JCGZ_24686 [Jatropha curcas] |
| 18 |
Hb_000665_110 |
0.2846004693 |
- |
- |
Ribosomal protein S11-beta [Theobroma cacao] |
| 19 |
Hb_000562_060 |
0.285388923 |
- |
- |
PREDICTED: uncharacterized protein LOC105169741 [Sesamum indicum] |
| 20 |
Hb_000097_010 |
0.2855088938 |
- |
- |
- |