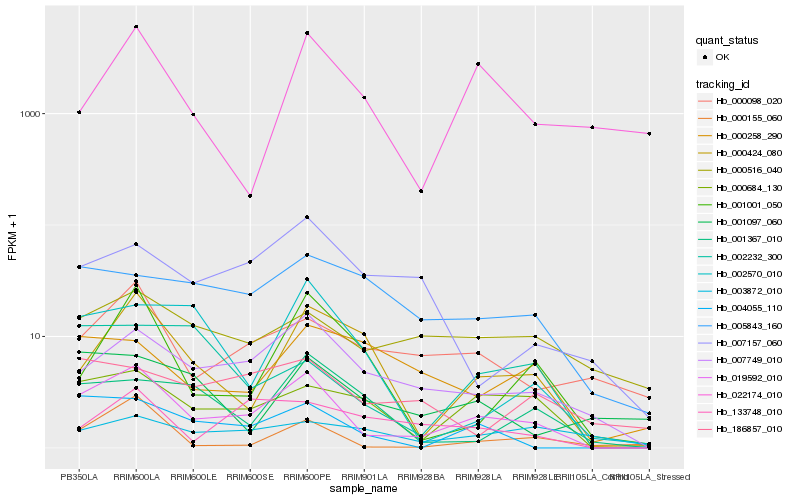
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_019592_010 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: TMV resistance protein N-like [Pyrus x bretschneideri] |
| 2 |
Hb_007749_010 |
0.2179320067 |
- |
- |
- |
| 3 |
Hb_000684_130 |
0.2320808439 |
- |
- |
- |
| 4 |
Hb_004055_110 |
0.24670985 |
- |
- |
PREDICTED: uncharacterized protein LOC105631442 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000424_080 |
0.2592958091 |
- |
- |
PREDICTED: endoglucanase 16 [Jatropha curcas] |
| 6 |
Hb_000098_020 |
0.2641118243 |
transcription factor |
TF Family: GRAS |
GRAS family transcription factor family protein [Theobroma cacao] |
| 7 |
Hb_000155_060 |
0.265695109 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_001001_050 |
0.2803394384 |
- |
- |
CXE protein [Hevea brasiliensis] |
| 9 |
Hb_000516_040 |
0.2823505695 |
- |
- |
PREDICTED: uncharacterized protein LOC105649502 isoform X2 [Jatropha curcas] |
| 10 |
Hb_005843_160 |
0.2882359229 |
- |
- |
PREDICTED: uncharacterized protein LOC105112024 [Populus euphratica] |
| 11 |
Hb_002570_010 |
0.2933721883 |
- |
- |
hypothetical protein JCGZ_25329 [Jatropha curcas] |
| 12 |
Hb_001367_010 |
0.2936002869 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 13 |
Hb_133748_010 |
0.2944647003 |
- |
- |
- |
| 14 |
Hb_002232_300 |
0.2975823516 |
- |
- |
Squalene monooxygenase, putative [Ricinus communis] |
| 15 |
Hb_007157_060 |
0.2991724193 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 16 |
Hb_022174_010 |
0.3019982498 |
- |
- |
elicitor-responsive protein [Hevea brasiliensis] |
| 17 |
Hb_186857_010 |
0.3031487697 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 18 |
Hb_000258_290 |
0.3036740378 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP15 [Jatropha curcas] |
| 19 |
Hb_001097_060 |
0.3046338313 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 8 [Jatropha curcas] |
| 20 |
Hb_003872_010 |
0.3072520634 |
- |
- |
hypothetical protein PRUPE_ppa013215mg [Prunus persica] |