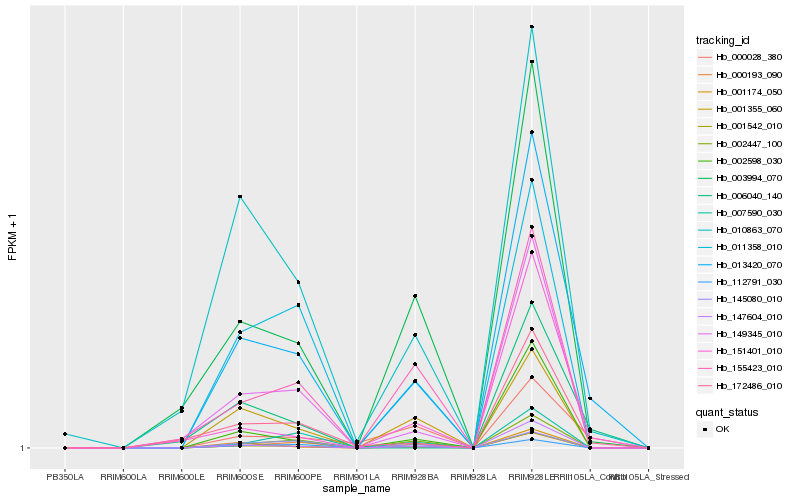
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013420_070 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_I01852 [Eucalyptus grandis] |
| 2 |
Hb_001542_010 |
0.1747012806 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 3 |
Hb_010863_070 |
0.1853588098 |
- |
- |
PREDICTED: glutaredoxin-C11 [Jatropha curcas] |
| 4 |
Hb_172486_010 |
0.1944818784 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 5 |
Hb_000028_380 |
0.1961935488 |
- |
- |
PREDICTED: uncharacterized protein LOC104212668 [Nicotiana sylvestris] |
| 6 |
Hb_002447_100 |
0.1980322042 |
- |
- |
hypothetical protein POPTR_0009s06260g [Populus trichocarpa] |
| 7 |
Hb_003994_070 |
0.1981528107 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 8 |
Hb_149345_010 |
0.1995151267 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 9 |
Hb_011358_010 |
0.2041117476 |
- |
- |
- |
| 10 |
Hb_001355_060 |
0.206488267 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 11 |
Hb_002598_030 |
0.2085920996 |
- |
- |
- |
| 12 |
Hb_112791_030 |
0.2151713642 |
- |
- |
Uncharacterized protein TCM_027940 [Theobroma cacao] |
| 13 |
Hb_151401_010 |
0.2169255918 |
- |
- |
- |
| 14 |
Hb_147604_010 |
0.2185046701 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 15 |
Hb_000193_090 |
0.2186350534 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 16 |
Hb_006040_140 |
0.2195741433 |
- |
- |
hypothetical protein NitaMp105 [Nicotiana tabacum] |
| 17 |
Hb_145080_010 |
0.2199756748 |
- |
- |
PREDICTED: uncharacterized protein LOC105643750 [Jatropha curcas] |
| 18 |
Hb_001174_050 |
0.2235230831 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_155423_010 |
0.2258992644 |
- |
- |
- |
| 20 |
Hb_007590_030 |
0.2276691138 |
- |
- |
PREDICTED: iridoid synthase [Jatropha curcas] |