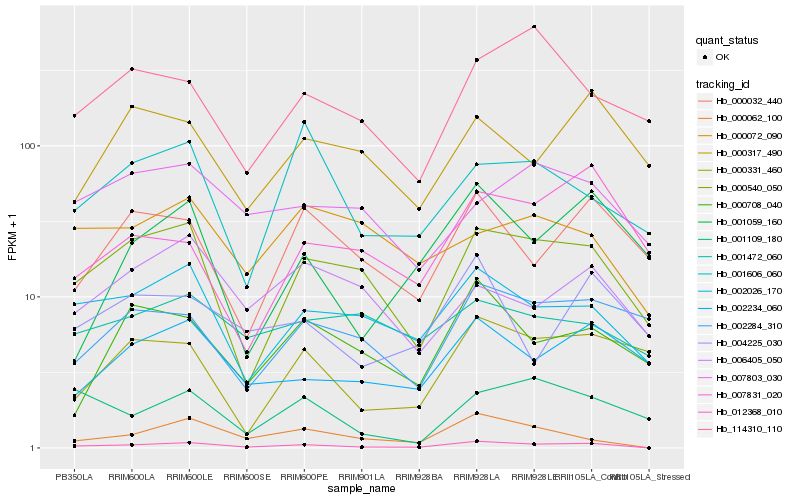
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012368_010 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At5g02620-like [Jatropha curcas] |
| 2 |
Hb_000062_100 |
0.1768361433 |
- |
- |
- |
| 3 |
Hb_000540_050 |
0.1882284123 |
- |
- |
PREDICTED: 125 kDa kinesin-related protein-like [Jatropha curcas] |
| 4 |
Hb_002026_170 |
0.1993764634 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 5 |
Hb_002234_060 |
0.2389492375 |
- |
- |
F-box protein, atfbl3, putative [Ricinus communis] |
| 6 |
Hb_000032_440 |
0.2411357674 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_000708_040 |
0.2416764177 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001606_060 |
0.2478756899 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001109_180 |
0.2490748034 |
- |
- |
PREDICTED: protein BREVIS RADIX-like [Populus euphratica] |
| 10 |
Hb_004225_030 |
0.2515109117 |
- |
- |
hypothetical protein F383_15788 [Gossypium arboreum] |
| 11 |
Hb_000331_460 |
0.2525078457 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 12 |
Hb_002284_310 |
0.2528909654 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001059_160 |
0.2544856909 |
- |
- |
hexose transporter 1 [Hevea brasiliensis] |
| 14 |
Hb_006405_050 |
0.2553341657 |
- |
- |
PREDICTED: stomatal closure-related actin-binding protein 2-like [Jatropha curcas] |
| 15 |
Hb_000072_090 |
0.2564755045 |
- |
- |
PREDICTED: SEC14 cytosolic factor [Jatropha curcas] |
| 16 |
Hb_114310_110 |
0.2565429952 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 17 |
Hb_007803_030 |
0.2588902439 |
- |
- |
PREDICTED: uncharacterized protein LOC105632791 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000317_490 |
0.2597200902 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 19 |
Hb_001472_060 |
0.262202493 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 8 isoform X1 [Jatropha curcas] |
| 20 |
Hb_007831_020 |
0.2655959014 |
- |
- |
conserved hypothetical protein [Ricinus communis] |