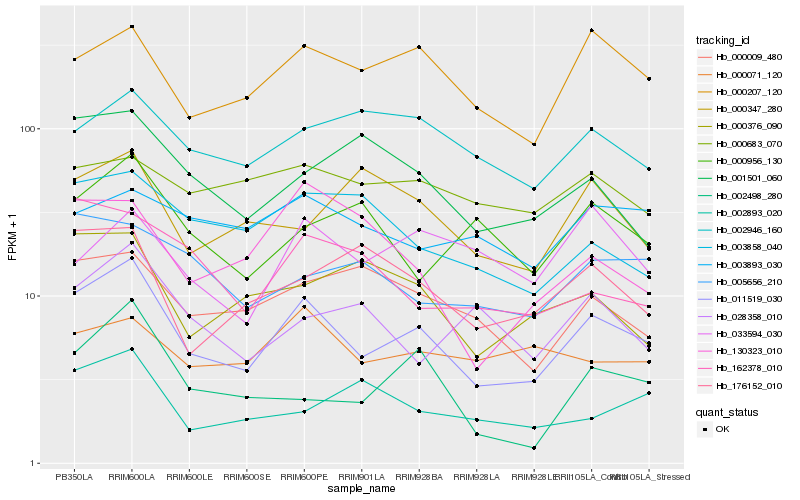
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011519_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein At3g17950 [Jatropha curcas] |
| 2 |
Hb_000207_120 |
0.1514032558 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.3 [Jatropha curcas] |
| 3 |
Hb_002498_280 |
0.1731040804 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 13C [Jatropha curcas] |
| 4 |
Hb_000009_480 |
0.1763435654 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At5g66560 [Jatropha curcas] |
| 5 |
Hb_003858_040 |
0.1800920583 |
- |
- |
hypothetical protein POPTR_0007s14590g [Populus trichocarpa] |
| 6 |
Hb_176152_010 |
0.180558685 |
- |
- |
Myosin heavy chain, fast skeletal muscle, embryonic, putative [Ricinus communis] |
| 7 |
Hb_000376_090 |
0.1819661275 |
- |
- |
Myosin heavy chain, fast skeletal muscle, embryonic, putative [Ricinus communis] |
| 8 |
Hb_005656_210 |
0.1823101927 |
- |
- |
PREDICTED: uncharacterized protein LOC105637440 [Jatropha curcas] |
| 9 |
Hb_002946_160 |
0.18242517 |
- |
- |
PREDICTED: uricase-2 isozyme 2 [Jatropha curcas] |
| 10 |
Hb_001501_060 |
0.1832058693 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_162378_010 |
0.1863977856 |
- |
- |
hypothetical protein JCGZ_20480 [Jatropha curcas] |
| 12 |
Hb_000071_120 |
0.1878437562 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 4 [Jatropha curcas] |
| 13 |
Hb_000956_130 |
0.1910067815 |
- |
- |
hypothetical protein POPTR_0002s12620g [Populus trichocarpa] |
| 14 |
Hb_028358_010 |
0.1912310651 |
- |
- |
PREDICTED: serine/threonine-protein kinase CTR1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_130323_010 |
0.1917382115 |
- |
- |
Myosin heavy chain, fast skeletal muscle, embryonic, putative [Ricinus communis] |
| 16 |
Hb_000683_070 |
0.1936080303 |
- |
- |
d-3-phosphoglycerate dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_003893_030 |
0.1939390618 |
- |
- |
Protein yrdA, putative [Ricinus communis] |
| 18 |
Hb_000347_280 |
0.1946550853 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 7 member B4-like [Populus euphratica] |
| 19 |
Hb_002893_020 |
0.1947566344 |
- |
- |
hypothetical protein JCGZ_21319 [Jatropha curcas] |
| 20 |
Hb_033594_030 |
0.1948339355 |
- |
- |
PREDICTED: putative glycosyltransferase 7 [Jatropha curcas] |