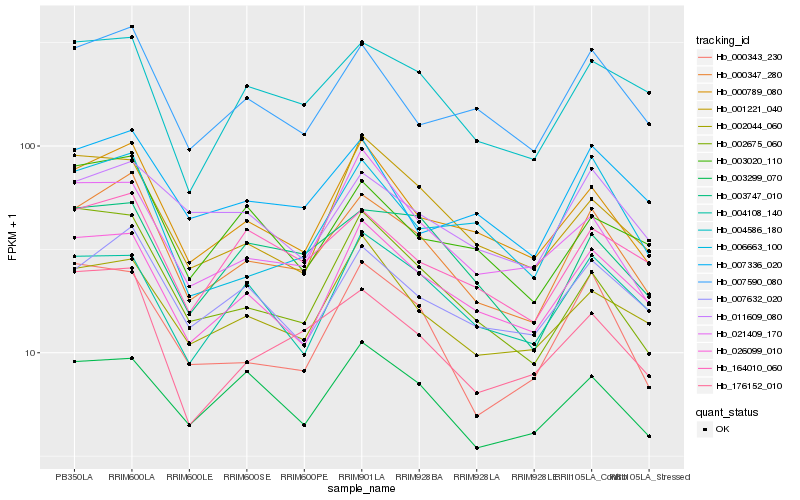
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_280 |
0.0 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 7 member B4-like [Populus euphratica] |
| 2 |
Hb_000789_080 |
0.0749919978 |
- |
- |
PREDICTED: arginine--tRNA ligase, cytoplasmic-like isoform X1 [Jatropha curcas] |
| 3 |
Hb_007632_020 |
0.0821216656 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 17 [Jatropha curcas] |
| 4 |
Hb_007590_080 |
0.0859664786 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP19-4 [Malus domestica] |
| 5 |
Hb_026099_010 |
0.0873855672 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 6 |
Hb_004586_180 |
0.0909495831 |
- |
- |
hypothetical protein JCGZ_15866 [Jatropha curcas] |
| 7 |
Hb_164010_060 |
0.0914938534 |
- |
- |
PREDICTED: uncharacterized protein LOC105633115 [Jatropha curcas] |
| 8 |
Hb_003747_010 |
0.0922655754 |
- |
- |
hypothetical protein JCGZ_24080 [Jatropha curcas] |
| 9 |
Hb_176152_010 |
0.0929989464 |
- |
- |
Myosin heavy chain, fast skeletal muscle, embryonic, putative [Ricinus communis] |
| 10 |
Hb_002044_060 |
0.0939779288 |
- |
- |
elongation factor ts, putative [Ricinus communis] |
| 11 |
Hb_000343_230 |
0.0979363012 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase PASTICCINO1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_011609_080 |
0.1004327743 |
- |
- |
uv excision repair protein rad23, putative [Ricinus communis] |
| 13 |
Hb_003299_070 |
0.1007990196 |
- |
- |
AMP deaminase [Theobroma cacao] |
| 14 |
Hb_002675_060 |
0.1008488379 |
- |
- |
PREDICTED: uncharacterized protein LOC105634954 isoform X2 [Jatropha curcas] |
| 15 |
Hb_007336_020 |
0.1010426549 |
- |
- |
Mannose-P-dolichol utilization defect 1 protein, putative [Ricinus communis] |
| 16 |
Hb_003020_110 |
0.1026178974 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 isoform X2 [Populus euphratica] |
| 17 |
Hb_001221_040 |
0.1033940878 |
- |
- |
PREDICTED: nucleolar protein 56-like [Jatropha curcas] |
| 18 |
Hb_006663_100 |
0.1040635241 |
- |
- |
hypothetical protein JCGZ_06980 [Jatropha curcas] |
| 19 |
Hb_021409_170 |
0.1050037913 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 20 |
Hb_004108_140 |
0.1051695255 |
- |
- |
PREDICTED: HUA2-like protein 3 isoform X3 [Jatropha curcas] |