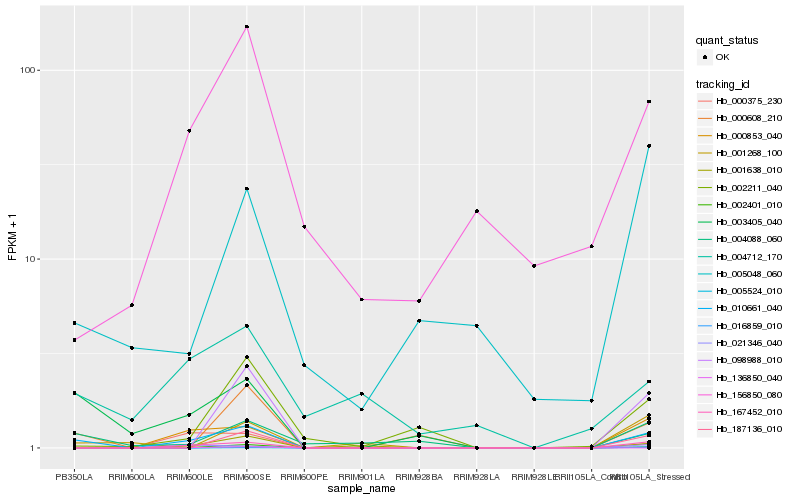
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010661_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0021s00470g [Populus trichocarpa] |
| 2 |
Hb_167452_010 |
0.2920364302 |
- |
- |
hypothetical protein JCGZ_03682 [Jatropha curcas] |
| 3 |
Hb_000853_040 |
0.3258923885 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_004088_060 |
0.3461071372 |
- |
- |
hypothetical protein POPTR_0012s01875g [Populus trichocarpa] |
| 5 |
Hb_005524_010 |
0.3559814118 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 6 |
Hb_000608_210 |
0.3607138878 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like protein FASCIATED EAR2 [Gossypium raimondii] |
| 7 |
Hb_003405_040 |
0.3613305594 |
- |
- |
PREDICTED: uncharacterized protein LOC105121978 [Populus euphratica] |
| 8 |
Hb_002401_010 |
0.3711086151 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 9 |
Hb_001638_010 |
0.3717789793 |
- |
- |
- |
| 10 |
Hb_021346_040 |
0.3795897565 |
- |
- |
hypothetical protein JCGZ_26751 [Jatropha curcas] |
| 11 |
Hb_156850_080 |
0.3911813404 |
- |
- |
PREDICTED: uncharacterized protein LOC105635247 [Jatropha curcas] |
| 12 |
Hb_002211_040 |
0.3922153635 |
- |
- |
PREDICTED: subtilisin-like protease SBT3.5 [Jatropha curcas] |
| 13 |
Hb_187136_010 |
0.3929707698 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 14 |
Hb_098988_010 |
0.3937688078 |
- |
- |
hypothetical protein JCGZ_18539 [Jatropha curcas] |
| 15 |
Hb_001268_100 |
0.3965737911 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein KNUCKLES [Jatropha curcas] |
| 16 |
Hb_016859_010 |
0.3976981281 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 17 |
Hb_136850_040 |
0.4017787664 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_005048_060 |
0.4026011563 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004712_170 |
0.4042557658 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 [Jatropha curcas] |
| 20 |
Hb_000375_230 |
0.4053904422 |
- |
- |
Chain A, Anomalous Substructure Of Hydroxynitrile Lyase |