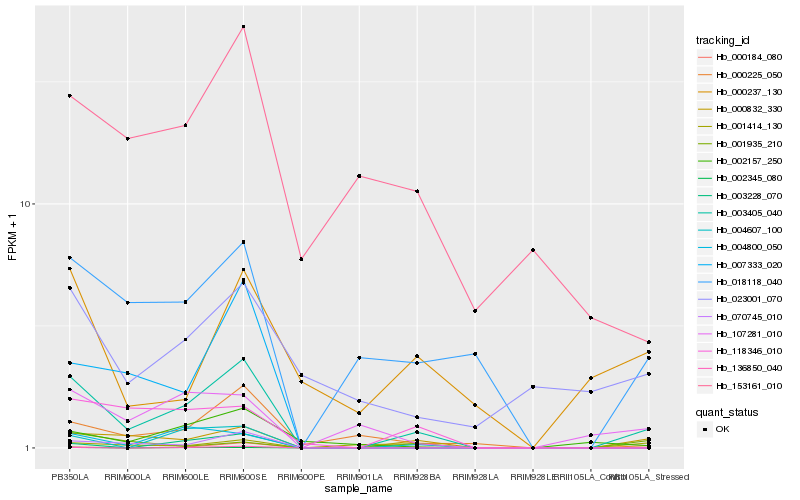
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003405_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105121978 [Populus euphratica] |
| 2 |
Hb_000832_330 |
0.2196786313 |
- |
- |
PREDICTED: uncharacterized protein LOC102661627 [Glycine max] |
| 3 |
Hb_070745_010 |
0.2900153747 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase A-like [Populus euphratica] |
| 4 |
Hb_018118_040 |
0.2930148675 |
- |
- |
- |
| 5 |
Hb_000225_050 |
0.2996739662 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 6 |
Hb_004607_100 |
0.2999683541 |
- |
- |
PREDICTED: uncharacterized protein LOC103445866 [Malus domestica] |
| 7 |
Hb_007333_020 |
0.3057568957 |
- |
- |
- |
| 8 |
Hb_118346_010 |
0.3154406353 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1 [Jatropha curcas] |
| 9 |
Hb_107281_010 |
0.3177003729 |
- |
- |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_002157_250 |
0.3277282632 |
- |
- |
Pectinesterase-4 precursor, putative [Ricinus communis] |
| 11 |
Hb_002345_080 |
0.329149757 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_136850_040 |
0.3332219251 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 13 |
Hb_004800_050 |
0.3337857228 |
- |
- |
PREDICTED: uncharacterized protein LOC105643859 [Jatropha curcas] |
| 14 |
Hb_000184_080 |
0.3363565198 |
- |
- |
contains similarity to reverse transcriptase (Pfam: rvt.hmm, score 19.29) [Arabidopsis thaliana] |
| 15 |
Hb_003228_070 |
0.3417104227 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001414_130 |
0.3463072502 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 17 |
Hb_153161_010 |
0.3496304635 |
- |
- |
phospholipid-transporting atpase, putative [Ricinus communis] |
| 18 |
Hb_001935_210 |
0.3551878103 |
- |
- |
- |
| 19 |
Hb_023001_070 |
0.3585334727 |
- |
- |
PREDICTED: dnaJ protein ERDJ3A-like [Eucalyptus grandis] |
| 20 |
Hb_000237_130 |
0.360696138 |
- |
- |
- |