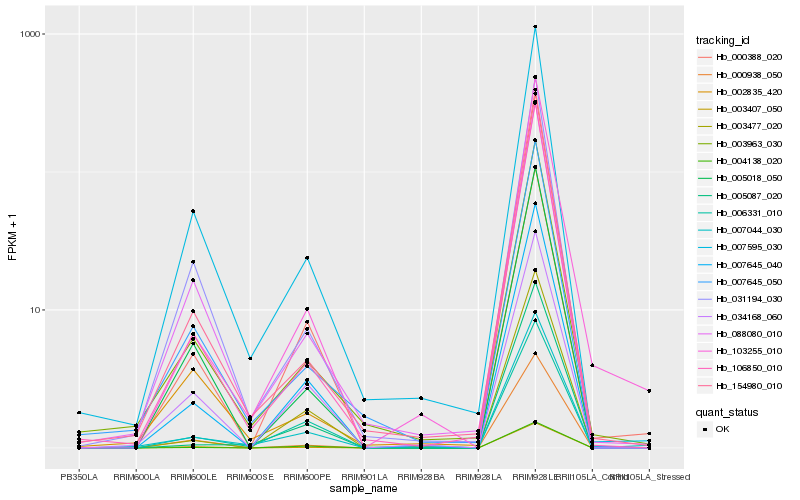
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007044_030 |
0.0 |
- |
- |
Os06g0273400 [Oryza sativa Japonica Group] |
| 2 |
Hb_007645_040 |
0.0829017915 |
- |
- |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Tachigali myrmecophila] |
| 3 |
Hb_003963_030 |
0.0875931753 |
- |
- |
psbB [Jatropha curcas] |
| 4 |
Hb_103255_010 |
0.0905999034 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 5 |
Hb_007595_030 |
0.0910688012 |
- |
- |
psbA gene product (chloroplast) [Silene noctiflora] |
| 6 |
Hb_034168_060 |
0.0921432356 |
- |
- |
ribulose-1-5-bisphosphate carboxylase/oxygenase large subunit, partial (chloroplast) [Cucumis argenteus] |
| 7 |
Hb_000388_020 |
0.0976061557 |
- |
- |
PREDICTED: MLP-like protein 43 [Fragaria vesca subsp. vesca] |
| 8 |
Hb_007645_050 |
0.0983127444 |
- |
- |
acetyl-CoA carboxylase beta subunit [Hevea brasiliensis] |
| 9 |
Hb_088080_010 |
0.0990960658 |
- |
- |
photosystem I P700 chlorophyll a apoprotein A1 [Matelea biflora] |
| 10 |
Hb_004138_020 |
0.1000954697 |
- |
- |
PREDICTED: cucumisin-like [Populus euphratica] |
| 11 |
Hb_005087_020 |
0.1011001474 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_006331_010 |
0.1020369713 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RKS1, partial [Populus euphratica] |
| 13 |
Hb_003407_050 |
0.1029945261 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 14 |
Hb_154980_010 |
0.1035422134 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 15 |
Hb_031194_030 |
0.1045111614 |
- |
- |
ribulose 1,5-bisphosphate carboxylase/oxygenase [Stackhousia minima] |
| 16 |
Hb_003477_020 |
0.1054572013 |
desease resistance |
Gene Name: AAA_29 |
PREDICTED: ABC transporter G family member 15-like [Populus euphratica] |
| 17 |
Hb_106850_010 |
0.1056115261 |
- |
- |
photosystem II protein D2 [Lactuca sativa] |
| 18 |
Hb_002835_420 |
0.1059567141 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 19 |
Hb_005018_050 |
0.1119916515 |
- |
- |
PREDICTED: licodione synthase-like [Jatropha curcas] |
| 20 |
Hb_000938_050 |
0.1123833984 |
- |
- |
hypothetical protein JCGZ_09640 [Jatropha curcas] |